Please be patient as the page loads
|
FREM3_HUMAN
|
||||||
| SwissProt Accessions | P0C091 | Gene names | FREM3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FREM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.871556 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
FREM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.858746 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
FREM2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987862 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SZK8, Q4QQG1, Q5H9N8, Q5T6Q1, Q6N057, Q6ZSB4, Q7Z305, Q7Z341 | Gene names | FREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog). | |||||
|
FREM2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986497 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6NVD0, Q4W2Q5, Q5H8C0, Q811G9, Q8C4G5, Q8CD46 | Gene names | Frem2, Nv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog) (NV domain-containing protein 1). | |||||
|
FREM3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P0C091 | Gene names | FREM3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor. | |||||
|
FREM3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994776 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H8B9 | Gene names | Frem3, Nv2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor (NV domain- containing protein 2). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 1.0077e-184 (rank : 7) | NC score | 0.644857 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 5.01279e-176 (rank : 8) | NC score | 0.638296 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
CSPG4_HUMAN
|
||||||
| θ value | 1.72781e-51 (rank : 9) | NC score | 0.721350 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
CSPG4_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 10) | NC score | 0.709507 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
NAC1_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 11) | NC score | 0.380023 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70414 | Gene names | Slc8a1, Ncx | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
NAC1_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 12) | NC score | 0.386870 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P32418, O95849, Q9UBL8, Q9UDN1, Q9UDN2, Q9UKX6 | Gene names | SLC8A1, CNC, NCX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
GPR98_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 13) | NC score | 0.301539 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
NAC3_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 14) | NC score | 0.369406 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P57103, Q8IUE9, Q8IUF0, Q8NFI7 | Gene names | SLC8A3, NCX3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 3 precursor (Na(+)/Ca(2+)-exchange protein 3). | |||||
|
NAC2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 15) | NC score | 0.342188 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPR5 | Gene names | SLC8A2, KIAA1087, NCX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 2 precursor (Na(+)/Ca(2+)-exchange protein 2). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.288885 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 17) | NC score | 0.092849 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.027909 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RSMN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.045298 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.045298 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.033212 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
PCDG3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.015327 (rank : 72) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDGA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.015438 (rank : 71) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
SPB6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.005963 (rank : 89) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35237, Q96J44 | Gene names | SERPINB6, PI6, PTI | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Cytoplasmic antiproteinase) (CAP) (Protease inhibitor 6) (PI-6). | |||||
|
P4HA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.015851 (rank : 70) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15460, Q8WWN0 | Gene names | P4HA2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.023630 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
DSC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.011208 (rank : 74) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.009999 (rank : 75) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
SYT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.009188 (rank : 79) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
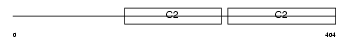
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.009128 (rank : 80) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
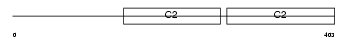
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
ZNF38_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | -0.001422 (rank : 96) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17036, P13683, Q9NNX8, Q9NXJ1, Q9UC15, Q9UC16 | Gene names | ZNF38, KOX25, ZNF3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 38 (Zinc finger protein KOX25) (Zinc finger protein HF.12) (Zinc finger protein 3) (HZF3.1). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.009818 (rank : 76) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PCDH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.012460 (rank : 73) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.003954 (rank : 92) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
OPLA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.023117 (rank : 68) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14841, Q75W65, Q9Y4Q0 | Gene names | OPLAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
OPLA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.023172 (rank : 67) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K010, Q8R3K2 | Gene names | Oplah | |||
|
Domain Architecture |

|
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
RENT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.008109 (rank : 82) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.005893 (rank : 90) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.008070 (rank : 83) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.009345 (rank : 78) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.009428 (rank : 77) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
RSMB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.031192 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |
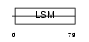
|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.006453 (rank : 88) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.006549 (rank : 87) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | -0.000271 (rank : 94) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
CAD23_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.029639 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022315 (rank : 69) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.000594 (rank : 93) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
GPR87_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | -0.001064 (rank : 95) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY21, Q96JZ8, Q9BXC2 | Gene names | GPR87, GPR95 | |||
|
Domain Architecture |
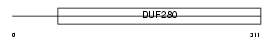
|
|||||
| Description | Probable G-protein coupled receptor 87 (G-protein coupled receptor 95). | |||||
|
IGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.008489 (rank : 81) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01344, P78449, Q14299, Q9UC68, Q9UC69 | Gene names | IGF2 | |||
|
Domain Architecture |
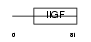
|
|||||
| Description | Insulin-like growth factor II precursor (IGF-II) (Somatomedin A) [Contains: Insulin-like growth factor II Ala-25 Del]. | |||||
|
MEF2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.005061 (rank : 91) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |
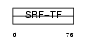
|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
RSMB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.028330 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |
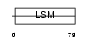
|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006698 (rank : 85) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
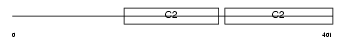
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.006678 (rank : 86) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
TNR16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.032384 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
ZN346_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.007489 (rank : 84) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0B7 | Gene names | Znf346, Jaz, Zfp346 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.090420 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.085016 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CHRD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.090382 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X0, O95254, Q6UW83, Q9H2D3, Q9H2W8, Q9H2W9, Q9P0Z2, Q9P0Z3, Q9P0Z4, Q9P0Z5 | Gene names | CHRD | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
CHRD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.086009 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
CRDL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.071269 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU40, Q9Y3H7 | Gene names | CHRDL1, NRLN1 | |||
|
Domain Architecture |
|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.064569 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920C1, Q924K0, Q9EPZ9 | Gene names | Chrdl1, Ng1, Nrln1 | |||
|
Domain Architecture |
|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.071260 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WN34, Q6WN30, Q6WN31, Q6WN32, Q7Z5J3 | Gene names | CHRDL2, BNF1, CHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor (Chordin-related protein 2) (Breast tumor novel factor 1) (BNF-1). | |||||
|
CRDL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.066770 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.084697 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.057930 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.061774 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.052113 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.058260 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.065839 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051591 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR410_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.055936 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.065821 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.052703 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.052113 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.081812 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.084156 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.056525 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.082855 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.080370 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.068497 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.060644 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050482 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059058 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052573 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.062830 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.079634 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
PCSK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.061247 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
REG3G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051487 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
RSPO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.055086 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
RSPO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.054710 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
RSPO3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051800 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXY4, Q5VTV4, Q96K87 | Gene names | RSPO3, PWTSR, THSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (hRspo3) (Thrombospondin type-1 domain-containing protein 2) (Protein with TSP type-1 repeat) (hPWTSR). | |||||
|
RSPO3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051187 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2TJ95, Q3SYI9, Q5R2V4, Q8BVW2, Q9CSB2 | Gene names | Rspo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (Cysteine-rich and single thrombospondin domain-containing protein 1) (Cristin-1) (Nucleopondin) (Cabriolet). | |||||
|
RSPO4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.062282 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
RSPO4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.062271 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050510 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FREM3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P0C091 | Gene names | FREM3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor. | |||||
|
FREM3_MOUSE
|
||||||
| NC score | 0.994776 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5H8B9 | Gene names | Frem3, Nv2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 3 precursor (NV domain- containing protein 2). | |||||
|
FREM2_HUMAN
|
||||||
| NC score | 0.987862 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SZK8, Q4QQG1, Q5H9N8, Q5T6Q1, Q6N057, Q6ZSB4, Q7Z305, Q7Z341 | Gene names | FREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog). | |||||
|
FREM2_MOUSE
|
||||||
| NC score | 0.986497 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6NVD0, Q4W2Q5, Q5H8C0, Q811G9, Q8C4G5, Q8CD46 | Gene names | Frem2, Nv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog) (NV domain-containing protein 1). | |||||
|
FREM1_HUMAN
|
||||||
| NC score | 0.871556 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
FREM1_MOUSE
|
||||||
| NC score | 0.858746 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
CSPG4_HUMAN
|
||||||
| NC score | 0.721350 (rank : 7) | θ value | 1.72781e-51 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
CSPG4_MOUSE
|
||||||
| NC score | 0.709507 (rank : 8) | θ value | 1.72781e-51 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.644857 (rank : 9) | θ value | 1.0077e-184 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.638296 (rank : 10) | θ value | 5.01279e-176 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
NAC1_HUMAN
|
||||||
| NC score | 0.386870 (rank : 11) | θ value | 8.67504e-11 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P32418, O95849, Q9UBL8, Q9UDN1, Q9UDN2, Q9UKX6 | Gene names | SLC8A1, CNC, NCX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
NAC1_MOUSE
|
||||||
| NC score | 0.380023 (rank : 12) | θ value | 2.98157e-11 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70414 | Gene names | Slc8a1, Ncx | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
NAC3_HUMAN
|
||||||
| NC score | 0.369406 (rank : 13) | θ value | 1.63604e-09 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P57103, Q8IUE9, Q8IUF0, Q8NFI7 | Gene names | SLC8A3, NCX3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 3 precursor (Na(+)/Ca(2+)-exchange protein 3). | |||||
|
NAC2_HUMAN
|
||||||
| NC score | 0.342188 (rank : 14) | θ value | 4.0297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPR5 | Gene names | SLC8A2, KIAA1087, NCX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 2 precursor (Na(+)/Ca(2+)-exchange protein 2). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.301539 (rank : 15) | θ value | 7.34386e-10 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.288885 (rank : 16) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.092849 (rank : 17) | θ value | 0.000158464 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.090420 (rank : 18) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
CHRD_HUMAN
|
||||||
| NC score | 0.090382 (rank : 19) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X0, O95254, Q6UW83, Q9H2D3, Q9H2W8, Q9H2W9, Q9P0Z2, Q9P0Z3, Q9P0Z4, Q9P0Z5 | Gene names | CHRD | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
CHRD_MOUSE
|
||||||
| NC score | 0.086009 (rank : 20) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0E2 | Gene names | Chrd | |||
|
Domain Architecture |

|
|||||
| Description | Chordin precursor. | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.085016 (rank : 21) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.084697 (rank : 22) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.084156 (rank : 23) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.082855 (rank : 24) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.081812 (rank : 25) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.080370 (rank : 26) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.079634 (rank : 27) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
CRDL1_HUMAN
|
||||||
| NC score | 0.071269 (rank : 28) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU40, Q9Y3H7 | Gene names | CHRDL1, NRLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
CRDL2_HUMAN
|
||||||
| NC score | 0.071260 (rank : 29) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WN34, Q6WN30, Q6WN31, Q6WN32, Q7Z5J3 | Gene names | CHRDL2, BNF1, CHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor (Chordin-related protein 2) (Breast tumor novel factor 1) (BNF-1). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.068497 (rank : 30) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
CRDL2_MOUSE
|
||||||
| NC score | 0.066770 (rank : 31) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.065839 (rank : 32) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.065821 (rank : 33) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
CRDL1_MOUSE
|
||||||
| NC score | 0.064569 (rank : 34) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920C1, Q924K0, Q9EPZ9 | Gene names | Chrdl1, Ng1, Nrln1 | |||
|
Domain Architecture |

|
|||||
| Description | Chordin-like protein 1 precursor (Neuralin-1) (Ventroptin) (Neurogenesin-1). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.062830 (rank : 35) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
RSPO4_HUMAN
|
||||||
| NC score | 0.062282 (rank : 36) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
RSPO4_MOUSE
|
||||||
| NC score | 0.062271 (rank : 37) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.061774 (rank : 38) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
PCSK6_HUMAN
|
||||||
| NC score | 0.061247 (rank : 39) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.060644 (rank : 40) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.059058 (rank : 41) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.058260 (rank : 42) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.057930 (rank : 43) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.056525 (rank : 44) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.055936 (rank : 45) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
RSPO2_HUMAN
|
||||||
| NC score | 0.055086 (rank : 46) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
RSPO2_MOUSE
|
||||||
| NC score | 0.054710 (rank : 47) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.052703 (rank : 48) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.052573 (rank : 49) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.052113 (rank : 50) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.052113 (rank : 51) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
RSPO3_HUMAN
|
||||||
| NC score | 0.051800 (rank : 52) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXY4, Q5VTV4, Q96K87 | Gene names | RSPO3, PWTSR, THSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (hRspo3) (Thrombospondin type-1 domain-containing protein 2) (Protein with TSP type-1 repeat) (hPWTSR). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.051591 (rank : 53) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
REG3G_MOUSE
|
||||||
| NC score | 0.051487 (rank : 54) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
RSPO3_MOUSE
|
||||||
| NC score | 0.051187 (rank : 55) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2TJ95, Q3SYI9, Q5R2V4, Q8BVW2, Q9CSB2 | Gene names | Rspo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (Cysteine-rich and single thrombospondin domain-containing protein 1) (Cristin-1) (Nucleopondin) (Cabriolet). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.050510 (rank : 56) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.050482 (rank : 57) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.045298 (rank : 58) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.045298 (rank : 59) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.033212 (rank : 60) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.032384 (rank : 61) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
RSMB_MOUSE
|
||||||
| NC score | 0.031192 (rank : 62) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
CAD23_MOUSE
|
||||||
| NC score | 0.029639 (rank : 63) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
RSMB_HUMAN
|
||||||
| NC score | 0.028330 (rank : 64) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.027909 (rank : 65) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | 0.023630 (rank : 66) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
OPLA_MOUSE
|
||||||
| NC score | 0.023172 (rank : 67) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K010, Q8R3K2 | Gene names | Oplah | |||
|
Domain Architecture |

|
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
OPLA_HUMAN
|
||||||
| NC score | 0.023117 (rank : 68) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14841, Q75W65, Q9Y4Q0 | Gene names | OPLAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.022315 (rank : 69) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
P4HA2_HUMAN
|
||||||
| NC score | 0.015851 (rank : 70) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15460, Q8WWN0 | Gene names | P4HA2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
PCDGA_HUMAN
|
||||||
| NC score | 0.015438 (rank : 71) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
PCDG3_HUMAN
|
||||||
| NC score | 0.015327 (rank : 72) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDH1_HUMAN
|
||||||
| NC score | 0.012460 (rank : 73) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
DSC1_HUMAN
|
||||||
| NC score | 0.011208 (rank : 74) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.009999 (rank : 75) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.009818 (rank : 76) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.009428 (rank : 77) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.009345 (rank : 78) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.009188 (rank : 79) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.009128 (rank : 80) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
IGF2_HUMAN
|
||||||
| NC score | 0.008489 (rank : 81) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01344, P78449, Q14299, Q9UC68, Q9UC69 | Gene names | IGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor II precursor (IGF-II) (Somatomedin A) [Contains: Insulin-like growth factor II Ala-25 Del]. | |||||
|
RENT1_HUMAN
|
||||||
| NC score | 0.008109 (rank : 82) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92900, O00239, O43343, Q86Z25, Q92842 | Gene names | UPF1, KIAA0221, RENT1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (hUpf1). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.008070 (rank : 83) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
ZN346_MOUSE
|
||||||
| NC score | 0.007489 (rank : 84) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0B7 | Gene names | Znf346, Jaz, Zfp346 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.006698 (rank : 85) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.006678 (rank : 86) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.006549 (rank : 87) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.006453 (rank : 88) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
SPB6_HUMAN
|
||||||
| NC score | 0.005963 (rank : 89) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35237, Q96J44 | Gene names | SERPINB6, PI6, PTI | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B6 (Placental thrombin inhibitor) (Cytoplasmic antiproteinase) (CAP) (Protease inhibitor 6) (PI-6). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.005893 (rank : 90) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
MEF2A_HUMAN
|
||||||
| NC score | 0.005061 (rank : 91) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.003954 (rank : 92) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.000594 (rank : 93) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | -0.000271 (rank : 94) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
GPR87_HUMAN
|
||||||
| NC score | -0.001064 (rank : 95) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY21, Q96JZ8, Q9BXC2 | Gene names | GPR87, GPR95 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 87 (G-protein coupled receptor 95). | |||||
|
ZNF38_HUMAN
|
||||||
| NC score | -0.001422 (rank : 96) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17036, P13683, Q9NNX8, Q9NXJ1, Q9UC15, Q9UC16 | Gene names | ZNF38, KOX25, ZNF3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 38 (Zinc finger protein KOX25) (Zinc finger protein HF.12) (Zinc finger protein 3) (HZF3.1). | |||||