Please be patient as the page loads
|
RNF25_HUMAN
|
||||||
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |
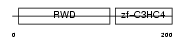
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RNF25_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 163 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
RNF25_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.949209 (rank : 2) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |
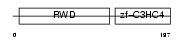
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
E2AK4_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 3) | NC score | 0.118018 (rank : 36) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
E2AK4_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 4) | NC score | 0.111160 (rank : 37) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
RWDD1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 5) | NC score | 0.353589 (rank : 3) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H446, Q9Y313, Q9Y6B3 | Gene names | RWDD1 | |||
|
Domain Architecture |
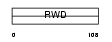
|
|||||
| Description | RWD domain-containing protein 1. | |||||
|
RWDD1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 6) | NC score | 0.351800 (rank : 4) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |
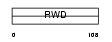
|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
RNF24_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.236802 (rank : 5) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGI1 | Gene names | Rnf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 24. | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.086331 (rank : 42) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.207321 (rank : 9) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
RNF24_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.226366 (rank : 6) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y225, Q9UMH1 | Gene names | RNF24 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 24. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.093867 (rank : 39) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PJA1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.197466 (rank : 12) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.201880 (rank : 10) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.071007 (rank : 60) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.094274 (rank : 38) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TF3C1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.063826 (rank : 67) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.088872 (rank : 41) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.071364 (rank : 57) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.058950 (rank : 74) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
RN122_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.217903 (rank : 8) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN122_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.218567 (rank : 7) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.069770 (rank : 62) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
RNF12_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.195643 (rank : 14) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.080561 (rank : 44) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN364_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.182885 (rank : 16) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.157783 (rank : 25) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.030077 (rank : 130) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
RNF38_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.196195 (rank : 13) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H0F5, Q8N0Y0 | Gene names | RNF38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 38. | |||||
|
RNF38_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.194423 (rank : 15) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BI21, Q6P5B1 | Gene names | Rnf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 38. | |||||
|
RNF6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.200758 (rank : 11) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
ZN364_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.179954 (rank : 17) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.061482 (rank : 71) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CU006_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.084509 (rank : 43) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99M03, Q9JLH4 | Gene names | ORF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf6 homolog. | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.036311 (rank : 122) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.057748 (rank : 77) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.028414 (rank : 136) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.053118 (rank : 91) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.021429 (rank : 152) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.043044 (rank : 112) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.056274 (rank : 81) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.073248 (rank : 50) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.044909 (rank : 109) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.058571 (rank : 75) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RN139_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.154868 (rank : 28) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WU17, O75485, Q7LDL3 | Gene names | RNF139, TRC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139 (Translocation in renal carcinoma on chromosome 8). | |||||
|
RNF13_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.174128 (rank : 21) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 13. | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.071190 (rank : 58) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
GOLI_MOUSE
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.173780 (rank : 22) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.076281 (rank : 47) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RN165_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.178707 (rank : 18) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
RNF13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.170570 (rank : 24) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43567 | Gene names | RNF13, RZF | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 13. | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.040578 (rank : 116) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
GOLI_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.171262 (rank : 23) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.046940 (rank : 104) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.020922 (rank : 153) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
RN139_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.148474 (rank : 31) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TMV1, Q8BZU9 | Gene names | Rnf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.029391 (rank : 132) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TMCC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.036827 (rank : 120) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.072183 (rank : 53) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.028879 (rank : 134) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
RN103_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.138355 (rank : 34) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00237, Q8IVB9 | Gene names | RNF103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103 homolog) (Zfp-103) (KF-1) (hKF-1). | |||||
|
RN103_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.139270 (rank : 33) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R1W3, O08670, O08883 | Gene names | Rnf103, Zfp103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103) (Zfp-103) (KF-1) (mKF-1). | |||||
|
TMCC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.035060 (rank : 123) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.029265 (rank : 133) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ECM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.045419 (rank : 108) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.025398 (rank : 141) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
INVS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.017937 (rank : 166) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.062892 (rank : 69) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.063540 (rank : 68) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.072269 (rank : 52) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.052841 (rank : 92) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.027348 (rank : 137) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FBLI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.020408 (rank : 155) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
G6PE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.024188 (rank : 142) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95479, Q4TT33, Q66I35, Q68DT3 | Gene names | H6PD, GDH | |||
|
Domain Architecture |

|
|||||
| Description | GDH/6PGL endoplasmic bifunctional protein precursor [Includes: Glucose 1-dehydrogenase (EC 1.1.1.47) (Hexose-6-phosphate dehydrogenase); 6- phosphogluconolactonase (EC 3.1.1.31) (6PGL)]. | |||||
|
GP152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.021676 (rank : 150) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.091880 (rank : 40) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.143215 (rank : 32) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.017429 (rank : 168) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
M3K3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.013361 (rank : 172) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99759 | Gene names | MAP3K3, MAPKKK3, MEKK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.056288 (rank : 80) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RN167_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.157434 (rank : 26) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H6Y7, Q6XYE0, Q8NDC1, Q9Y3V1 | Gene names | RNF167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RNF11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.178199 (rank : 19) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y3C5 | Gene names | RNF11 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 11 (Sid 1669). | |||||
|
RNF11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.177693 (rank : 20) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYK7, Q9EQI1 | Gene names | Rnf11, N4wbp2, Sid1669 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 11 (NEDD4 WW domain-binding protein 2) (Sid 1669). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.047802 (rank : 102) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.062068 (rank : 70) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.030674 (rank : 129) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
YBOX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.021663 (rank : 151) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.053573 (rank : 88) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
COJA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.044225 (rank : 110) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.018576 (rank : 163) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.066816 (rank : 65) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.051999 (rank : 93) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.023250 (rank : 145) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.045421 (rank : 107) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.055296 (rank : 86) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RN126_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.150863 (rank : 30) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.005403 (rank : 186) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.032359 (rank : 126) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.018029 (rank : 165) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.011728 (rank : 177) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.009560 (rank : 181) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.016574 (rank : 170) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.053269 (rank : 90) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
CN004_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.037095 (rank : 119) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.053511 (rank : 89) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.043711 (rank : 111) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CP007_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.023291 (rank : 144) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2B5 | Gene names | C16orf7, ATPBL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 (Protein ATP-BL). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.014196 (rank : 171) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.020719 (rank : 154) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.029547 (rank : 131) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.049176 (rank : 100) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PABP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.021859 (rank : 149) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.049512 (rank : 99) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCY1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.022466 (rank : 147) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811Q9, Q3UEW0, Q80Y63, Q811Q8, Q8BKD2, Q8C085 | Gene names | Pcyt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
RN133_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.155148 (rank : 27) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVZ7, Q8N7G7 | Gene names | RNF133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 133. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.048392 (rank : 101) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.047624 (rank : 103) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.042957 (rank : 113) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.050052 (rank : 97) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.045549 (rank : 105) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.050970 (rank : 95) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.049923 (rank : 98) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.027340 (rank : 138) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.020364 (rank : 156) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FOXP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.007068 (rank : 183) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H334, Q9H332, Q9H333, Q9P0R1 | Gene names | FOXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P1. | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.031354 (rank : 127) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.011008 (rank : 178) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INVO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.022510 (rank : 146) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.027237 (rank : 139) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.007328 (rank : 182) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.006570 (rank : 185) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.019338 (rank : 158) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.039993 (rank : 117) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.016722 (rank : 169) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SDK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.007044 (rank : 184) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
TS13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.036625 (rank : 121) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.013198 (rank : 173) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
WBP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.020118 (rank : 157) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.030682 (rank : 128) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ZSCA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | -0.002446 (rank : 187) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z7L9, Q6ZQY9, Q9NWU4 | Gene names | ZSCAN2, ZFP29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29 homolog) (Zfp-29). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.018757 (rank : 162) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.038947 (rank : 118) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.028668 (rank : 135) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BUB1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.012488 (rank : 174) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
CF057_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.022389 (rank : 148) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VUM1 | Gene names | C6orf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0369 protein C6orf57 precursor. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.023571 (rank : 143) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.041172 (rank : 115) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.042715 (rank : 114) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.012483 (rank : 175) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
HRG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.019177 (rank : 160) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
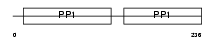
Domain Architecture |
|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
ICB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.012459 (rank : 176) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TEJ8, O60560, Q5TEJ1, Q5TEJ9, Q5TEK1, Q68DP4, Q9BYB6, Q9NS90 | Gene names | ICB1, C1orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein (Protein ICB-1). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.026751 (rank : 140) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
NDUB9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.017436 (rank : 167) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
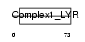
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
NUD12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.010177 (rank : 179) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
PCY1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.018529 (rank : 164) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5K3, O60621, Q86XC9 | Gene names | PCYT1B, CCTB | |||
|
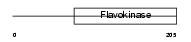
Domain Architecture |
|
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
RN167_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.151917 (rank : 29) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XF4, Q3U4S5 | Gene names | Rnf167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RWDD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.034506 (rank : 124) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3V2, Q6FID3, Q9BX35 | Gene names | RWDD3 | |||
|
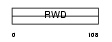
Domain Architecture |
|
|||||
| Description | RWD domain-containing protein 3. | |||||
|
RWDD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.045501 (rank : 106) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPR1 | Gene names | Rwdd4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RWD domain-containing protein 4A. | |||||
|
SYN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.033040 (rank : 125) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |
|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
TNNT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.009719 (rank : 180) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
UBR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.019149 (rank : 161) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
UBR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.019213 (rank : 159) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.050548 (rank : 96) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ZSCA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | -0.002735 (rank : 188) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
AMFR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.075126 (rank : 49) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
AMFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.075727 (rank : 48) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R049, Q8K008, Q99LH5 | Gene names | Amfr | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor (EC 6.3.2.19) (AMF receptor). | |||||
|
CBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.067045 (rank : 64) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CU006_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051299 (rank : 94) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57060 | Gene names | C21orf6 | |||
|
Domain Architecture |
|
|||||
| Description | Protein C21orf6. | |||||
|
DZIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.071433 (rank : 56) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.071458 (rank : 55) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.055444 (rank : 85) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.055596 (rank : 83) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.055501 (rank : 84) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.072096 (rank : 54) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053593 (rank : 87) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.060564 (rank : 72) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RBX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.056448 (rank : 79) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF6, Q9BXN8, Q9Y5M7 | Gene names | RNF7, RBX2, ROC2, SAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING-box protein 2 (Rbx2) (RING finger protein 7) (Regulator of cullins 2) (CKII beta-binding protein 1) (CKBBP1) (Sensitive to apoptosis gene protein). | |||||
|
RBX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.056587 (rank : 78) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTZ1, Q3UKF8 | Gene names | Rnf7, Rbx2, Sag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING-box protein 2 (Rbx2) (RING finger protein 7) (Sensitive to apoptosis gene protein). | |||||
|
RN126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.135862 (rank : 35) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN141_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.070186 (rank : 61) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVD5, Q9NZB4 | Gene names | RNF141, ZNF230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
RN141_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.071027 (rank : 59) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99MB7, Q9D040, Q9D5C4, Q9D9L0 | Gene names | Rnf141, Znf230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
RN175_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.056238 (rank : 82) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N4F7 | Gene names | RNF175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 175. | |||||
|
RNF32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060145 (rank : 73) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
RNF32_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.058166 (rank : 76) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
SYHH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.072688 (rank : 51) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49590 | Gene names | HARSL, HARSR, HO3 | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase homolog (EC 6.1.1.21) (Histidine--tRNA ligase homolog) (HisRS). | |||||
|
SYH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.076765 (rank : 46) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12081 | Gene names | HARS, HRS | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
SYH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.077714 (rank : 45) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61035 | Gene names | Hars | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
ZN363_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.066115 (rank : 66) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PM5, Q96PR5 | Gene names | RCHY1, ARNIP, CHIMP, PIRH2, ZNF363 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger and CHY zinc finger domain-containing protein 1 (Zinc finger protein 363) (CH-rich-interacting match with PLAG1) (Androgen receptor N-terminal-interacting protein) (p53 induced RING-H2 protein) (hPirh2). | |||||
|
ZN363_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.068534 (rank : 63) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CR50, Q920H0 | Gene names | Rchy1, Arnip, Chimp, Zfp363, Znf363 | |||
|
Domain Architecture |
|
|||||
| Description | RING finger and CHY zinc finger domain-containing protein 1 (Zinc finger protein 363) (CH-rich-interacting match with PLAG1) (Androgen receptor N-terminal-interacting protein). | |||||
|
RNF25_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 163 | |
| SwissProt Accessions | Q96BH1, Q9H874 | Gene names | RNF25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-). | |||||
|
RNF25_MOUSE
|
||||||
| NC score | 0.949209 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
RWDD1_HUMAN
|
||||||
| NC score | 0.353589 (rank : 3) | θ value | 1.43324e-05 (rank : 5) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H446, Q9Y313, Q9Y6B3 | Gene names | RWDD1 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 1. | |||||
|
RWDD1_MOUSE
|
||||||
| NC score | 0.351800 (rank : 4) | θ value | 1.87187e-05 (rank : 6) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
RNF24_MOUSE
|
||||||
| NC score | 0.236802 (rank : 5) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGI1 | Gene names | Rnf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 24. | |||||
|
RNF24_HUMAN
|
||||||
| NC score | 0.226366 (rank : 6) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y225, Q9UMH1 | Gene names | RNF24 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 24. | |||||
|
RN122_MOUSE
|
||||||
| NC score | 0.218567 (rank : 7) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN122_HUMAN
|
||||||
| NC score | 0.217903 (rank : 8) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.207321 (rank : 9) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.201880 (rank : 10) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
RNF6_HUMAN
|
||||||
| NC score | 0.200758 (rank : 11) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
PJA1_HUMAN
|
||||||
| NC score | 0.197466 (rank : 12) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
RNF38_HUMAN
|
||||||
| NC score | 0.196195 (rank : 13) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H0F5, Q8N0Y0 | Gene names | RNF38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 38. | |||||
|
RNF12_HUMAN
|
||||||
| NC score | 0.195643 (rank : 14) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
RNF38_MOUSE
|
||||||
| NC score | 0.194423 (rank : 15) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BI21, Q6P5B1 | Gene names | Rnf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 38. | |||||
|
ZN364_HUMAN
|
||||||
| NC score | 0.182885 (rank : 16) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
ZN364_MOUSE
|
||||||
| NC score | 0.179954 (rank : 17) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.178707 (rank : 18) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
RNF11_HUMAN
|
||||||
| NC score | 0.178199 (rank : 19) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y3C5 | Gene names | RNF11 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 11 (Sid 1669). | |||||
|
RNF11_MOUSE
|
||||||
| NC score | 0.177693 (rank : 20) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYK7, Q9EQI1 | Gene names | Rnf11, N4wbp2, Sid1669 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 11 (NEDD4 WW domain-binding protein 2) (Sid 1669). | |||||
|
RNF13_MOUSE
|
||||||
| NC score | 0.174128 (rank : 21) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 13. | |||||
|
GOLI_MOUSE
|
||||||
| NC score | 0.173780 (rank : 22) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VEM1, Q80VL7, Q9QZQ6 | Gene names | Rnf130, G1rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130) (G1-related zinc finger protein). | |||||
|
GOLI_HUMAN
|
||||||
| NC score | 0.171262 (rank : 23) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86XS8, Q641P9, Q6P177, Q7L2U2, Q9P0J9 | Gene names | RNF130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Goliath homolog precursor (RING finger protein 130). | |||||
|
RNF13_HUMAN
|
||||||
| NC score | 0.170570 (rank : 24) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43567 | Gene names | RNF13, RZF | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 13. | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.157783 (rank : 25) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
RN167_HUMAN
|
||||||
| NC score | 0.157434 (rank : 26) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H6Y7, Q6XYE0, Q8NDC1, Q9Y3V1 | Gene names | RNF167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RN133_HUMAN
|
||||||
| NC score | 0.155148 (rank : 27) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVZ7, Q8N7G7 | Gene names | RNF133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 133. | |||||
|
RN139_HUMAN
|
||||||
| NC score | 0.154868 (rank : 28) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WU17, O75485, Q7LDL3 | Gene names | RNF139, TRC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139 (Translocation in renal carcinoma on chromosome 8). | |||||
|
RN167_MOUSE
|
||||||
| NC score | 0.151917 (rank : 29) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XF4, Q3U4S5 | Gene names | Rnf167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 167 precursor. | |||||
|
RN126_MOUSE
|
||||||
| NC score | 0.150863 (rank : 30) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN139_MOUSE
|
||||||
| NC score | 0.148474 (rank : 31) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7TMV1, Q8BZU9 | Gene names | Rnf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 139. | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.143215 (rank : 32) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
RN103_MOUSE
|
||||||
| NC score | 0.139270 (rank : 33) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R1W3, O08670, O08883 | Gene names | Rnf103, Zfp103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103) (Zfp-103) (KF-1) (mKF-1). | |||||
|
RN103_HUMAN
|
||||||
| NC score | 0.138355 (rank : 34) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00237, Q8IVB9 | Gene names | RNF103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103 homolog) (Zfp-103) (KF-1) (hKF-1). | |||||
|
RN126_HUMAN
|
||||||
| NC score | 0.135862 (rank : 35) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
E2AK4_MOUSE
|
||||||
| NC score | 0.118018 (rank : 36) | θ value | 1.133e-10 (rank : 3) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
E2AK4_HUMAN
|
||||||
| NC score | 0.111160 (rank : 37) | θ value | 6.21693e-09 (rank : 4) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.094274 (rank : 38) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.093867 (rank : 39) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.091880 (rank : 40) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.088872 (rank : 41) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.086331 (rank : 42) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
CU006_MOUSE
|
||||||
| NC score | 0.084509 (rank : 43) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99M03, Q9JLH4 | Gene names | ORF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf6 homolog. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.080561 (rank : 44) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SYH_MOUSE
|
||||||
| NC score | 0.077714 (rank : 45) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61035 | Gene names | Hars | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
SYH_HUMAN
|
||||||
| NC score | 0.076765 (rank : 46) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12081 | Gene names | HARS, HRS | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.076281 (rank : 47) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
AMFR_MOUSE
|
||||||
| NC score | 0.075727 (rank : 48) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R049, Q8K008, Q99LH5 | Gene names | Amfr | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor (EC 6.3.2.19) (AMF receptor). | |||||
|
AMFR2_HUMAN
|
||||||
| NC score | 0.075126 (rank : 49) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.073248 (rank : 50) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
SYHH_HUMAN
|
||||||
| NC score | 0.072688 (rank : 51) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49590 | Gene names | HARSL, HARSR, HO3 | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase homolog (EC 6.1.1.21) (Histidine--tRNA ligase homolog) (HisRS). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.072269 (rank : 52) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.072183 (rank : 53) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.072096 (rank : 54) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.071458 (rank : 55) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
DZIP3_HUMAN
|
||||||
| NC score | 0.071433 (rank : 56) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.071364 (rank : 57) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.071190 (rank : 58) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
RN141_MOUSE
|
||||||
| NC score | 0.071027 (rank : 59) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99MB7, Q9D040, Q9D5C4, Q9D9L0 | Gene names | Rnf141, Znf230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.071007 (rank : 60) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
RN141_HUMAN
|
||||||
| NC score | 0.070186 (rank : 61) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVD5, Q9NZB4 | Gene names | RNF141, ZNF230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 141 (Zinc finger protein 230). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.069770 (rank : 62) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ZN363_MOUSE
|
||||||
| NC score | 0.068534 (rank : 63) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CR50, Q920H0 | Gene names | Rchy1, Arnip, Chimp, Zfp363, Znf363 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and CHY zinc finger domain-containing protein 1 (Zinc finger protein 363) (CH-rich-interacting match with PLAG1) (Androgen receptor N-terminal-interacting protein). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.067045 (rank : 64) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.066816 (rank : 65) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ZN363_HUMAN
|
||||||
| NC score | 0.066115 (rank : 66) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PM5, Q96PR5 | Gene names | RCHY1, ARNIP, CHIMP, PIRH2, ZNF363 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and CHY zinc finger domain-containing protein 1 (Zinc finger protein 363) (CH-rich-interacting match with PLAG1) (Androgen receptor N-terminal-interacting protein) (p53 induced RING-H2 protein) (hPirh2). | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.063826 (rank : 67) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.063540 (rank : 68) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.062892 (rank : 69) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.062068 (rank : 70) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.061482 (rank : 71) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.060564 (rank : 72) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RNF32_HUMAN
|
||||||
| NC score | 0.060145 (rank : 73) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.058950 (rank : 74) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.058571 (rank : 75) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RNF32_MOUSE
|
||||||
| NC score | 0.058166 (rank : 76) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.057748 (rank : 77) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
RBX2_MOUSE
|
||||||
| NC score | 0.056587 (rank : 78) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTZ1, Q3UKF8 | Gene names | Rnf7, Rbx2, Sag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING-box protein 2 (Rbx2) (RING finger protein 7) (Sensitive to apoptosis gene protein). | |||||
|
RBX2_HUMAN
|
||||||
| NC score | 0.056448 (rank : 79) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF6, Q9BXN8, Q9Y5M7 | Gene names | RNF7, RBX2, ROC2, SAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING-box protein 2 (Rbx2) (RING finger protein 7) (Regulator of cullins 2) (CKII beta-binding protein 1) (CKBBP1) (Sensitive to apoptosis gene protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.056288 (rank : 80) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.056274 (rank : 81) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
RN175_HUMAN
|
||||||
| NC score | 0.056238 (rank : 82) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N4F7 | Gene names | RNF175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 175. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.055596 (rank : 83) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.055501 (rank : 84) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.055444 (rank : 85) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.055296 (rank : 86) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.053593 (rank : 87) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.053573 (rank : 88) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.053511 (rank : 89) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.053269 (rank : 90) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.053118 (rank : 91) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.052841 (rank : 92) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.051999 (rank : 93) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CU006_HUMAN
|
||||||
| NC score | 0.051299 (rank : 94) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57060 | Gene names | C21orf6 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf6. | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.050970 (rank : 95) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.050548 (rank : 96) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.050052 (rank : 97) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.049923 (rank : 98) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.049512 (rank : 99) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.049176 (rank : 100) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.048392 (rank : 101) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.047802 (rank : 102) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.047624 (rank : 103) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.046940 (rank : 104) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.045549 (rank : 105) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
RWDD4_MOUSE
|
||||||
| NC score | 0.045501 (rank : 106) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CPR1 | Gene names | Rwdd4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RWD domain-containing protein 4A. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.045421 (rank : 107) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ECM1_MOUSE
|
||||||
| NC score | 0.045419 (rank : 108) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.044909 (rank : 109) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.044225 (rank : 110) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.043711 (rank : 111) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.043044 (rank : 112) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.042957 (rank : 113) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.042715 (rank : 114) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.041172 (rank : 115) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.040578 (rank : 116) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.039993 (rank : 117) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.038947 (rank : 118) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.037095 (rank : 119) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
TMCC1_MOUSE
|
||||||
| NC score | 0.036827 (rank : 120) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.036625 (rank : 121) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.036311 (rank : 122) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
TMCC1_HUMAN
|
||||||
| NC score | 0.035060 (rank : 123) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
RWDD3_HUMAN
|
||||||
| NC score | 0.034506 (rank : 124) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3V2, Q6FID3, Q9BX35 | Gene names | RWDD3 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 3. | |||||
|
SYN3_HUMAN
|
||||||
| NC score | 0.033040 (rank : 125) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.032359 (rank : 126) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.031354 (rank : 127) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.030682 (rank : 128) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.030674 (rank : 129) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.030077 (rank : 130) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.029547 (rank : 131) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.029391 (rank : 132) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.029265 (rank : 133) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.028879 (rank : 134) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.028668 (rank : 135) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.028414 (rank : 136) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.027348 (rank : 137) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.027340 (rank : 138) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.027237 (rank : 139) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.026751 (rank : 140) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.025398 (rank : 141) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
G6PE_HUMAN
|
||||||
| NC score | 0.024188 (rank : 142) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95479, Q4TT33, Q66I35, Q68DT3 | Gene names | H6PD, GDH | |||
|
Domain Architecture |

|
|||||
| Description | GDH/6PGL endoplasmic bifunctional protein precursor [Includes: Glucose 1-dehydrogenase (EC 1.1.1.47) (Hexose-6-phosphate dehydrogenase); 6- phosphogluconolactonase (EC 3.1.1.31) (6PGL)]. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.023571 (rank : 143) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CP007_HUMAN
|
||||||
| NC score | 0.023291 (rank : 144) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2B5 | Gene names | C16orf7, ATPBL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 (Protein ATP-BL). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.023250 (rank : 145) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.022510 (rank : 146) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
PCY1B_MOUSE
|
||||||
| NC score | 0.022466 (rank : 147) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811Q9, Q3UEW0, Q80Y63, Q811Q8, Q8BKD2, Q8C085 | Gene names | Pcyt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
CF057_HUMAN
|
||||||
| NC score | 0.022389 (rank : 148) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VUM1 | Gene names | C6orf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0369 protein C6orf57 precursor. | |||||
|
PABP2_HUMAN
|
||||||
| NC score | 0.021859 (rank : 149) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.021676 (rank : 150) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
YBOX2_HUMAN
|
||||||
| NC score | 0.021663 (rank : 151) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.021429 (rank : 152) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.020922 (rank : 153) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.020719 (rank : 154) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
FBLI1_MOUSE
|
||||||
| NC score | 0.020408 (rank : 155) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.020364 (rank : 156) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
WBP4_MOUSE
|
||||||
| NC score | 0.020118 (rank : 157) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.019338 (rank : 158) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
UBR2_MOUSE
|
||||||
| NC score | 0.019213 (rank : 159) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
HRG_HUMAN
|
||||||
| NC score | 0.019177 (rank : 160) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |

|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
UBR2_HUMAN
|
||||||
| NC score | 0.019149 (rank : 161) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.018757 (rank : 162) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.018576 (rank : 163) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
PCY1B_HUMAN
|
||||||
| NC score | 0.018529 (rank : 164) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5K3, O60621, Q86XC9 | Gene names | PCYT1B, CCTB | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.018029 (rank : 165) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.017937 (rank : 166) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
NDUB9_HUMAN
|
||||||
| NC score | 0.017436 (rank : 167) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.017429 (rank : 168) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.016722 (rank : 169) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.016574 (rank : 170) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.014196 (rank : 171) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
M3K3_HUMAN
|
||||||
| NC score | 0.013361 (rank : 172) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99759 | Gene names | MAP3K3, MAPKKK3, MEKK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.013198 (rank : 173) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
BUB1B_HUMAN
|
||||||
| NC score | 0.012488 (rank : 174) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.012483 (rank : 175) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ICB1_HUMAN
|
||||||
| NC score | 0.012459 (rank : 176) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TEJ8, O60560, Q5TEJ1, Q5TEJ9, Q5TEK1, Q68DP4, Q9BYB6, Q9NS90 | Gene names | ICB1, C1orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein (Protein ICB-1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.011728 (rank : 177) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.011008 (rank : 178) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
NUD12_HUMAN
|
||||||
| NC score | 0.010177 (rank : 179) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
TNNT2_MOUSE
|
||||||
| NC score | 0.009719 (rank : 180) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.009560 (rank : 181) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.007328 (rank : 182) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
FOXP1_HUMAN
|
||||||
| NC score | 0.007068 (rank : 183) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H334, Q9H332, Q9H333, Q9P0R1 | Gene names | FOXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P1. | |||||
|
SDK1_HUMAN
|
||||||
| NC score | 0.007044 (rank : 184) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.006570 (rank : 185) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.005403 (rank : 186) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
ZSCA2_HUMAN
|
||||||
| NC score | -0.002446 (rank : 187) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z7L9, Q6ZQY9, Q9NWU4 | Gene names | ZSCAN2, ZFP29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29 homolog) (Zfp-29). | |||||
|
ZSCA2_MOUSE
|
||||||
| NC score | -0.002735 (rank : 188) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 163 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||