Please be patient as the page loads
|
UBE1L_HUMAN
|
||||||
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBE1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979326 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979274 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 4) | NC score | 0.749358 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
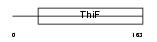
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 5) | NC score | 0.744313 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
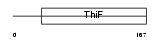
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 6) | NC score | 0.714179 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 7) | NC score | 0.724723 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 8) | NC score | 0.723858 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
ULE1B_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 9) | NC score | 0.706471 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULA1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 10) | NC score | 0.572999 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 11) | NC score | 0.562917 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
MOCS3_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 12) | NC score | 0.617097 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
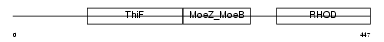
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
UE1D1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 13) | NC score | 0.396311 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 14) | NC score | 0.397457 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
ATG7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.125290 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
ATG7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.124242 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ERN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.005120 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76MJ5, Q6ZNC0 | Gene names | ERN2, IRE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase/endoribonuclease IRE2 precursor (Inositol-requiring protein 2) (hIRE2p) (IRE1b) (Ire1-beta) (Endoplasmic reticulum-to-nucleus signaling 2) [Includes: Serine/threonine-protein kinase (EC 2.7.11.1); Endoribonuclease (EC 3.1.26.-)]. | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.024871 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SLIK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.005738 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.009035 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
PA24D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.007743 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.008645 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
EDRF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.019569 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3B7T1, Q4G190, Q8IZ74, Q9Y3W4 | Gene names | EDRF1, C10orf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
EDRF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.019437 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
PIGZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.014772 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VD9, Q9H9G6 | Gene names | PIGZ, SMP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI mannosyltransferase 4 (EC 2.4.1.-) (GPI mannosyltransferase IV) (GPI-MT-IV) (Phosphatidylinositol-glycan biosynthesis class Z protein) (PIG-Z) (hSMP3). | |||||
|
UNC5D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.003439 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.979326 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.979274 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.749358 (rank : 4) | θ value | 2.35151e-24 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.744313 (rank : 5) | θ value | 5.23862e-24 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.724723 (rank : 6) | θ value | 4.43474e-23 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.723858 (rank : 7) | θ value | 5.79196e-23 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.714179 (rank : 8) | θ value | 1.99067e-23 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.706471 (rank : 9) | θ value | 3.51386e-20 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.617097 (rank : 10) | θ value | 2.52405e-10 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.572999 (rank : 11) | θ value | 7.09661e-13 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.562917 (rank : 12) | θ value | 1.02475e-11 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.397457 (rank : 13) | θ value | 1.87187e-05 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.396311 (rank : 14) | θ value | 4.92598e-06 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
ATG7_HUMAN
|
||||||
| NC score | 0.125290 (rank : 15) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95352, Q7L8L0, Q9BWP2, Q9UFH4 | Gene names | ATG7, APG7L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (hAGP7). | |||||
|
ATG7_MOUSE
|
||||||
| NC score | 0.124242 (rank : 16) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.024871 (rank : 17) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
EDRF1_HUMAN
|
||||||
| NC score | 0.019569 (rank : 18) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3B7T1, Q4G190, Q8IZ74, Q9Y3W4 | Gene names | EDRF1, C10orf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
EDRF1_MOUSE
|
||||||
| NC score | 0.019437 (rank : 19) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
PIGZ_HUMAN
|
||||||
| NC score | 0.014772 (rank : 20) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VD9, Q9H9G6 | Gene names | PIGZ, SMP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI mannosyltransferase 4 (EC 2.4.1.-) (GPI mannosyltransferase IV) (GPI-MT-IV) (Phosphatidylinositol-glycan biosynthesis class Z protein) (PIG-Z) (hSMP3). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.009035 (rank : 21) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.008645 (rank : 22) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
PA24D_HUMAN
|
||||||
| NC score | 0.007743 (rank : 23) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
SLIK5_HUMAN
|
||||||
| NC score | 0.005738 (rank : 24) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||
|
ERN2_HUMAN
|
||||||
| NC score | 0.005120 (rank : 25) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76MJ5, Q6ZNC0 | Gene names | ERN2, IRE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase/endoribonuclease IRE2 precursor (Inositol-requiring protein 2) (hIRE2p) (IRE1b) (Ire1-beta) (Endoplasmic reticulum-to-nucleus signaling 2) [Includes: Serine/threonine-protein kinase (EC 2.7.11.1); Endoribonuclease (EC 3.1.26.-)]. | |||||
|
UNC5D_MOUSE
|
||||||
| NC score | 0.003439 (rank : 26) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||