Please be patient as the page loads
|
UE1D1_MOUSE
|
||||||
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UE1D1_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
UE1D1_HUMAN
|
||||||
| θ value | 7.9845e-182 (rank : 2) | NC score | 0.996681 (rank : 2) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 3) | NC score | 0.485163 (rank : 4) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
MOCS3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 4) | NC score | 0.503647 (rank : 3) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |
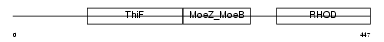
|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
UBE1L_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 5) | NC score | 0.396311 (rank : 8) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 6) | NC score | 0.414091 (rank : 6) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 7) | NC score | 0.412251 (rank : 7) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 8) | NC score | 0.381725 (rank : 9) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 9) | NC score | 0.377395 (rank : 10) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
ULE1B_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 10) | NC score | 0.428095 (rank : 5) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.278940 (rank : 11) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |
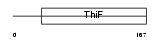
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.270224 (rank : 12) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |
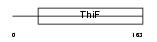
|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.004661 (rank : 19) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
ANR10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.006697 (rank : 17) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
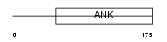
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
41_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.005398 (rank : 18) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.013281 (rank : 16) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
ATG7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.050225 (rank : 15) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
ULA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.148986 (rank : 14) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ULA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.151396 (rank : 13) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
UE1D1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VE47, Q9CYD6 | Gene names | Ube1dc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme). | |||||
|
UE1D1_HUMAN
|
||||||
| NC score | 0.996681 (rank : 2) | θ value | 7.9845e-182 (rank : 2) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZZ9, Q96ST1 | Gene names | UBE1DC1, UBA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-activating enzyme E1 domain-containing protein 1 (UFM1- activating enzyme) (Ubiquitin-activating enzyme 5) (ThiFP1). | |||||
|
MOCS3_HUMAN
|
||||||
| NC score | 0.503647 (rank : 3) | θ value | 2.61198e-07 (rank : 4) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95396 | Gene names | MOCS3 | |||
|
Domain Architecture |

|
|||||
| Description | Molybdenum cofactor synthesis protein 3 (Molybdopterin synthase sulfurylase) (MPT synthase sulfurylase). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.485163 (rank : 4) | θ value | 1.9326e-10 (rank : 3) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.428095 (rank : 5) | θ value | 7.1131e-05 (rank : 10) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.414091 (rank : 6) | θ value | 3.19293e-05 (rank : 6) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.412251 (rank : 7) | θ value | 4.1701e-05 (rank : 7) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBE1L_HUMAN
|
||||||
| NC score | 0.396311 (rank : 8) | θ value | 4.92598e-06 (rank : 5) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41226 | Gene names | UBE1L, UBE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 homolog (D8). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.381725 (rank : 9) | θ value | 4.1701e-05 (rank : 8) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UBE1_HUMAN
|
||||||
| NC score | 0.377395 (rank : 10) | θ value | 7.1131e-05 (rank : 9) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22314, Q96E13 | Gene names | UBE1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 (A1S9 protein). | |||||
|
ULE1A_MOUSE
|
||||||
| NC score | 0.278940 (rank : 11) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T2, Q3TWJ0, Q9CSW9 | Gene names | Uble1a, Aos1, Sae1, Sua1, Ubl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULE1A_HUMAN
|
||||||
| NC score | 0.270224 (rank : 12) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBE0, O95717, Q9P020 | Gene names | UBLE1A, AOS1, SAE1, SUA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1A (SUMO-1-activating enzyme subunit 1). | |||||
|
ULA1_MOUSE
|
||||||
| NC score | 0.151396 (rank : 13) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VBW6, Q8CFL6 | Gene names | Appbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1). | |||||
|
ULA1_HUMAN
|
||||||
| NC score | 0.148986 (rank : 14) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13564 | Gene names | APPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 regulatory subunit (Amyloid protein-binding protein 1) (Amyloid beta precursor protein-binding protein 1, 59 kDa) (APP-BP1) (Protooncogene protein 1) (HPP1). | |||||
|
ATG7_MOUSE
|
||||||
| NC score | 0.050225 (rank : 15) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D906, Q3TCD9, Q8K4Q5 | Gene names | Atg7, Apg7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 7 (APG7-like) (Ubiquitin-activating enzyme E1-like protein) (mAGP7). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.013281 (rank : 16) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.006697 (rank : 17) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
41_HUMAN
|
||||||
| NC score | 0.005398 (rank : 18) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.004661 (rank : 19) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||