Please be patient as the page loads
|
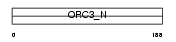
ORC3_HUMAN
|
||||||
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ORC3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
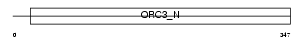
ORC3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981236 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 3) | NC score | 0.027427 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
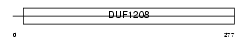
SHLB2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 4) | NC score | 0.054093 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR46, Q8WY61, Q96JH9 | Gene names | SH3GLB2, KIAA1848 | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain GRB2-like protein B2 (Endophilin-B2). | |||||
|
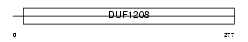
SHLB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 5) | NC score | 0.053908 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3V5, Q8K140, Q91ZI7 | Gene names | Sh3glb2, Kiaa1848 | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain GRB2-like protein B2 (Endophilin-B2). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.025980 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.022944 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.019033 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.023997 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.015320 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.021324 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.024892 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.018171 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
SYMPK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.049941 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.049696 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.019179 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.021189 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
HEAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.028906 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
SMRD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.020372 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6STE5, Q75MJ2, Q75MR8, Q92926, Q9BUH1 | Gene names | SMARCD3, BAF60C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C). | |||||
|
SMRD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.020375 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
APOA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.024492 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
ARL8A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.005670 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BM9 | Gene names | ARL8A, ARL10B, GIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 8A (ADP-ribosylation factor-like protein 10B) (Novel small G protein indispensable for equal chromosome segregation 2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.014861 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LRRK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.002395 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.015849 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.008789 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.017961 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.017064 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CBPE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.007081 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.018238 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
ECEL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.007689 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.013451 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
OLR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.015274 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
PPME1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.023456 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
PPME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.023200 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.018524 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.007882 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.008018 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.022378 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ORC3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.981236 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
SHLB2_HUMAN
|
||||||
| NC score | 0.054093 (rank : 3) | θ value | 0.365318 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR46, Q8WY61, Q96JH9 | Gene names | SH3GLB2, KIAA1848 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain GRB2-like protein B2 (Endophilin-B2). | |||||
|
SHLB2_MOUSE
|
||||||
| NC score | 0.053908 (rank : 4) | θ value | 0.365318 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3V5, Q8K140, Q91ZI7 | Gene names | Sh3glb2, Kiaa1848 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain GRB2-like protein B2 (Endophilin-B2). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.049941 (rank : 5) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.049696 (rank : 6) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
HEAT2_HUMAN
|
||||||
| NC score | 0.028906 (rank : 7) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.027427 (rank : 8) | θ value | 0.365318 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.025980 (rank : 9) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.024892 (rank : 10) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.024492 (rank : 11) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.023997 (rank : 12) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
PPME1_HUMAN
|
||||||
| NC score | 0.023456 (rank : 13) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
PPME1_MOUSE
|
||||||
| NC score | 0.023200 (rank : 14) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.022944 (rank : 15) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.022378 (rank : 16) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.021324 (rank : 17) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.021189 (rank : 18) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
SMRD3_MOUSE
|
||||||
| NC score | 0.020375 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
SMRD3_HUMAN
|
||||||
| NC score | 0.020372 (rank : 20) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6STE5, Q75MJ2, Q75MR8, Q92926, Q9BUH1 | Gene names | SMARCD3, BAF60C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.019179 (rank : 21) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.019033 (rank : 22) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.018524 (rank : 23) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.018238 (rank : 24) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.018171 (rank : 25) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.017961 (rank : 26) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.017064 (rank : 27) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.015849 (rank : 28) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.015320 (rank : 29) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
OLR1_MOUSE
|
||||||
| NC score | 0.015274 (rank : 30) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.014861 (rank : 31) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.013451 (rank : 32) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.008789 (rank : 33) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.008018 (rank : 34) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.007882 (rank : 35) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
ECEL1_MOUSE
|
||||||
| NC score | 0.007689 (rank : 36) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| NC score | 0.007081 (rank : 37) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
ARL8A_HUMAN
|
||||||
| NC score | 0.005670 (rank : 38) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BM9 | Gene names | ARL8A, ARL10B, GIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 8A (ADP-ribosylation factor-like protein 10B) (Novel small G protein indispensable for equal chromosome segregation 2). | |||||
|
LRRK2_HUMAN
|
||||||
| NC score | 0.002395 (rank : 39) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||