Please be patient as the page loads
|
SYMPK_HUMAN
|
||||||
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYMPK_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978265 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 3) | NC score | 0.058798 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.119356 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.079555 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RAF1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.039157 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04049 | Gene names | RAF1, RAF | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
RAF1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.039196 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.022671 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.074445 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
ARAF_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.036554 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
BRAF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.034021 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15056, Q13878, Q9Y6T3 | Gene names | BRAF, BRAF1, RAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | B-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (p94) (v-Raf murine sarcoma viral oncogene homolog B1). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.036552 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
NAB2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.101956 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.034392 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.037863 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.068837 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.027968 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.052333 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.022743 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.035134 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NAB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.089029 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.047828 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
ICB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.037608 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.034999 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.037723 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.035774 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.035832 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
PI3R4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.038596 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99570 | Gene names | PIK3R4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4) (PI3-kinase p150 subunit) (Phosphoinositide 3-kinase adaptor protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.022314 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.029093 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ORC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.062276 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.019229 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.037104 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.028899 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.030136 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.030488 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
VAV3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.019578 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.020431 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.039668 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
FBW1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.018405 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKB1, Q9P2S8, Q9P2S9, Q9Y4C6 | Gene names | FBXW11, BTRCP2, FBW1B, FBXW1B, KIAA0696 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 11 (F-box/WD repeat protein 1B) (F-box and WD repeats protein beta-TrCP2). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.027815 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ERR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.008298 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11474, Q14514 | Gene names | ESRRA, ERR1, ESRL1, NR3B1 | |||
|
Domain Architecture |
|
|||||
| Description | Steroid hormone receptor ERR1 (Estrogen-related receptor, alpha) (ERR- alpha) (Estrogen receptor-like 1). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.033290 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.028025 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SACS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.041878 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
ORC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.049941 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
STX18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.036530 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.021402 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
ANR33_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.019150 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXP5, Q8BXP4, Q8R1Z1 | Gene names | Ankrd33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
CA103_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.021037 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDD9, Q8C893 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf103 homolog. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.020805 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.027008 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.020274 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.067688 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.026950 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PIAS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.020494 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.033506 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.037879 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.010299 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.013298 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ZSCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.001432 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.012669 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.027150 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.017019 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
EXOSX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.015868 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
FBW1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.013615 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007291 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.039231 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.004338 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009514 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.011232 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.020989 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.022939 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SVIL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.020696 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012960 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.007655 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CH1B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.011336 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.011719 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
LRRK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.010367 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.026880 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.011087 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.005078 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NOL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.019838 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.018287 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PIAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016701 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.016859 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.010456 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.008483 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.014801 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
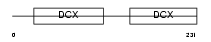
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.020843 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
IF39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.055904 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.978265 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.119356 (rank : 3) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
NAB2_HUMAN
|
||||||
| NC score | 0.101956 (rank : 4) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
NAB2_MOUSE
|
||||||
| NC score | 0.089029 (rank : 5) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.079555 (rank : 6) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.074445 (rank : 7) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.068837 (rank : 8) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.067688 (rank : 9) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.062276 (rank : 10) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.058798 (rank : 11) | θ value | 0.000461057 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.055904 (rank : 12) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.052333 (rank : 13) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
ORC3_HUMAN
|
||||||
| NC score | 0.049941 (rank : 14) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.047828 (rank : 15) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.041878 (rank : 16) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.039668 (rank : 17) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.039231 (rank : 18) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
RAF1_MOUSE
|
||||||
| NC score | 0.039196 (rank : 19) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
RAF1_HUMAN
|
||||||
| NC score | 0.039157 (rank : 20) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04049 | Gene names | RAF1, RAF | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
PI3R4_HUMAN
|
||||||
| NC score | 0.038596 (rank : 21) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99570 | Gene names | PIK3R4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4) (PI3-kinase p150 subunit) (Phosphoinositide 3-kinase adaptor protein). | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.037879 (rank : 22) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.037863 (rank : 23) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.037723 (rank : 24) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ICB1_MOUSE
|
||||||
| NC score | 0.037608 (rank : 25) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.037104 (rank : 26) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ARAF_MOUSE
|
||||||
| NC score | 0.036554 (rank : 27) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.036552 (rank : 28) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.036530 (rank : 29) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.035832 (rank : 30) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.035774 (rank : 31) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.035134 (rank : 32) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.034999 (rank : 33) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ARAF_HUMAN
|
||||||
| NC score | 0.034392 (rank : 34) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
BRAF1_HUMAN
|
||||||
| NC score | 0.034021 (rank : 35) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15056, Q13878, Q9Y6T3 | Gene names | BRAF, BRAF1, RAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | B-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (p94) (v-Raf murine sarcoma viral oncogene homolog B1). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.033506 (rank : 36) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.033290 (rank : 37) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.030488 (rank : 38) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.030136 (rank : 39) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029093 (rank : 40) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.028899 (rank : 41) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.028025 (rank : 42) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.027968 (rank : 43) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.027815 (rank : 44) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.027150 (rank : 45) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.027008 (rank : 46) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.026950 (rank : 47) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.026880 (rank : 48) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.022939 (rank : 49) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.022743 (rank : 50) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.022671 (rank : 51) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.022314 (rank : 52) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.021402 (rank : 53) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
CA103_MOUSE
|
||||||
| NC score | 0.021037 (rank : 54) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDD9, Q8C893 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf103 homolog. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.020989 (rank : 55) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.020843 (rank : 56) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.020805 (rank : 57) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
SVIL_HUMAN
|
||||||
| NC score | 0.020696 (rank : 58) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
PIAS1_HUMAN
|
||||||
| NC score | 0.020494 (rank : 59) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.020431 (rank : 60) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.020274 (rank : 61) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
NOL1_HUMAN
|
||||||
| NC score | 0.019838 (rank : 62) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.019578 (rank : 63) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.019229 (rank : 64) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
ANR33_MOUSE
|
||||||
| NC score | 0.019150 (rank : 65) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXP5, Q8BXP4, Q8R1Z1 | Gene names | Ankrd33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
FBW1B_HUMAN
|
||||||
| NC score | 0.018405 (rank : 66) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKB1, Q9P2S8, Q9P2S9, Q9Y4C6 | Gene names | FBXW11, BTRCP2, FBW1B, FBXW1B, KIAA0696 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 11 (F-box/WD repeat protein 1B) (F-box and WD repeats protein beta-TrCP2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.018287 (rank : 67) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.017019 (rank : 68) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.016859 (rank : 69) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PIAS1_MOUSE
|
||||||
| NC score | 0.016701 (rank : 70) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
EXOSX_HUMAN
|
||||||
| NC score | 0.015868 (rank : 71) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.014801 (rank : 72) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
FBW1A_HUMAN
|
||||||
| NC score | 0.013615 (rank : 73) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.013298 (rank : 74) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.012960 (rank : 75) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.012669 (rank : 76) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.011719 (rank : 77) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
CH1B1_MOUSE
|
||||||
| NC score | 0.011336 (rank : 78) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
PANK2_HUMAN
|
||||||
| NC score | 0.011232 (rank : 79) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.011087 (rank : 80) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.010456 (rank : 81) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
LRRK1_MOUSE
|
||||||
| NC score | 0.010367 (rank : 82) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.010299 (rank : 83) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.009514 (rank : 84) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.008483 (rank : 85) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
ERR1_HUMAN
|
||||||
| NC score | 0.008298 (rank : 86) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11474, Q14514 | Gene names | ESRRA, ERR1, ESRL1, NR3B1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR1 (Estrogen-related receptor, alpha) (ERR- alpha) (Estrogen receptor-like 1). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.007655 (rank : 87) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.007291 (rank : 88) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.005078 (rank : 89) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.004338 (rank : 90) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
ZSCA1_HUMAN
|
||||||
| NC score | 0.001432 (rank : 91) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||