Please be patient as the page loads
|
SYMPK_MOUSE
|
||||||
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYMPK_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.978265 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
IF39_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 3) | NC score | 0.085686 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.085395 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
RAF1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.039284 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04049 | Gene names | RAF1, RAF | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
RAF1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.039469 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.116144 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.079248 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
ARAF_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.036853 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.073642 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BRAF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.034010 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15056, Q13878, Q9Y6T3 | Gene names | BRAF, BRAF1, RAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | B-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (p94) (v-Raf murine sarcoma viral oncogene homolog B1). | |||||
|
NAB2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.098705 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.049261 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.035111 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.047772 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RBFAL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.064704 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3B9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ribosome-binding factor A, mitochondrial precursor. | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.034490 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.023408 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.043451 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NAB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.086280 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.010792 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.020547 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.039717 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.040828 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.066347 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
IRX6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.025158 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78412, Q7Z2K0 | Gene names | IRX6, IRX7, IRXB3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-6 (Iroquois homeobox protein 6) (Homeodomain protein IRXB3). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.011026 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.042423 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.050804 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.031480 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.006266 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.042362 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.048892 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
M4A12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.036085 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXJ0, Q8N6L4 | Gene names | MS4A12 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane-spanning 4-domains subfamily A member 12. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.012596 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
ORC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.061993 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PI3R4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.037693 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99570 | Gene names | PIK3R4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4) (PI3-kinase p150 subunit) (Phosphoinositide 3-kinase adaptor protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.029948 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.052379 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
PAXI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.018010 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.047149 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SACS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.046885 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SYLM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.035928 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
FBW1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.018549 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKB1, Q9P2S8, Q9P2S9, Q9Y4C6 | Gene names | FBXW11, BTRCP2, FBW1B, FBXW1B, KIAA0696 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 11 (F-box/WD repeat protein 1B) (F-box and WD repeats protein beta-TrCP2). | |||||
|
KIFC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.009495 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
PAXI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.017278 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.062512 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.015358 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.033226 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CLAT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.015577 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28329, Q16488, Q9BQ23, Q9BQ35, Q9BQE1 | Gene names | CHAT | |||
|
Domain Architecture |

|
|||||
| Description | Choline O-acetyltransferase (EC 2.3.1.6) (CHOACTase) (Choline acetylase) (ChAT). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.028026 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.028086 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
IPP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.030031 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCL8, Q3UD25, Q8C1A6 | Gene names | Ppp1r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase inhibitor 2 (IPP-2). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.042772 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.034412 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.033749 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.025786 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.024016 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.021304 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.019420 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
ORC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.049696 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.022664 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
STX18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.037605 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.033614 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.021563 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.020834 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.039657 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.041124 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PIAS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.020366 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
RAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.014160 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.034481 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.020589 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SYLM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.030390 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.015004 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.004831 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANR33_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.019667 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXP5, Q8BXP4, Q8R1Z1 | Gene names | Ankrd33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.024753 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.016919 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
FBW1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.013772 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009575 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.005568 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
M3K4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.010486 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6R4, Q92612 | Gene names | MAP3K4, KIAA0213, MAPKKK4, MEKK4, MTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4) (MAP three kinase 1). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.026380 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.006633 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.009925 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.022132 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.007700 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.050339 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006625 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.020232 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SVIL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.018416 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TNR21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.011200 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.020972 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013645 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.024203 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.013170 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.023657 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.012626 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.012877 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.014938 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LRRK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.011477 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.006006 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.034410 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PIAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.016584 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
PO2F2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005198 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.014689 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
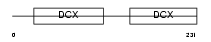
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010235 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.019809 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | -0.000717 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.978265 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.116144 (rank : 3) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
NAB2_HUMAN
|
||||||
| NC score | 0.098705 (rank : 4) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15742, O76006, Q14797 | Gene names | NAB2, MADER | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2) (Melanoma- associated delayed early response protein) (Protein MADER). | |||||
|
NAB2_MOUSE
|
||||||
| NC score | 0.086280 (rank : 5) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61127 | Gene names | Nab2 | |||
|
Domain Architecture |

|
|||||
| Description | NGFI-A-binding protein 2 (EGR-1-binding protein 2). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.085686 (rank : 6) | θ value | 0.00665767 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.085395 (rank : 7) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.079248 (rank : 8) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.073642 (rank : 9) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.066347 (rank : 10) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RBFAL_MOUSE
|
||||||
| NC score | 0.064704 (rank : 11) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3B9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ribosome-binding factor A, mitochondrial precursor. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.062512 (rank : 12) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.061993 (rank : 13) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.052379 (rank : 14) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.050804 (rank : 15) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050339 (rank : 16) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ORC3_HUMAN
|
||||||
| NC score | 0.049696 (rank : 17) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBD5, Q13565, Q6IUY7, Q9UG44, Q9UNT6 | Gene names | ORC3L, LATHEO | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.049261 (rank : 18) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.048892 (rank : 19) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.047772 (rank : 20) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.047149 (rank : 21) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.046885 (rank : 22) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.043451 (rank : 23) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.042772 (rank : 24) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.042423 (rank : 25) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.042362 (rank : 26) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.041124 (rank : 27) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.040828 (rank : 28) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.039717 (rank : 29) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.039657 (rank : 30) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
RAF1_MOUSE
|
||||||
| NC score | 0.039469 (rank : 31) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
RAF1_HUMAN
|
||||||
| NC score | 0.039284 (rank : 32) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04049 | Gene names | RAF1, RAF | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
PI3R4_HUMAN
|
||||||
| NC score | 0.037693 (rank : 33) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99570 | Gene names | PIK3R4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4) (PI3-kinase p150 subunit) (Phosphoinositide 3-kinase adaptor protein). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.037605 (rank : 34) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
ARAF_MOUSE
|
||||||
| NC score | 0.036853 (rank : 35) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
M4A12_HUMAN
|
||||||
| NC score | 0.036085 (rank : 36) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXJ0, Q8N6L4 | Gene names | MS4A12 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-spanning 4-domains subfamily A member 12. | |||||
|
SYLM_HUMAN
|
||||||
| NC score | 0.035928 (rank : 37) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.035111 (rank : 38) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
ARAF_HUMAN
|
||||||
| NC score | 0.034490 (rank : 39) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.034481 (rank : 40) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.034412 (rank : 41) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.034410 (rank : 42) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
BRAF1_HUMAN
|
||||||
| NC score | 0.034010 (rank : 43) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15056, Q13878, Q9Y6T3 | Gene names | BRAF, BRAF1, RAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | B-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (p94) (v-Raf murine sarcoma viral oncogene homolog B1). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.033749 (rank : 44) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.033614 (rank : 45) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.033226 (rank : 46) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.031480 (rank : 47) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SYLM_MOUSE
|
||||||
| NC score | 0.030390 (rank : 48) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
IPP2_MOUSE
|
||||||
| NC score | 0.030031 (rank : 49) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCL8, Q3UD25, Q8C1A6 | Gene names | Ppp1r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase inhibitor 2 (IPP-2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.029948 (rank : 50) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.028086 (rank : 51) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.028026 (rank : 52) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.026380 (rank : 53) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.025786 (rank : 54) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
IRX6_HUMAN
|
||||||
| NC score | 0.025158 (rank : 55) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78412, Q7Z2K0 | Gene names | IRX6, IRX7, IRXB3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-6 (Iroquois homeobox protein 6) (Homeodomain protein IRXB3). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.024753 (rank : 56) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.024203 (rank : 57) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.024016 (rank : 58) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.023657 (rank : 59) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.023408 (rank : 60) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.022664 (rank : 61) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.022132 (rank : 62) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.021563 (rank : 63) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.021304 (rank : 64) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.020972 (rank : 65) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.020834 (rank : 66) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.020589 (rank : 67) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.020547 (rank : 68) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
PIAS1_HUMAN
|
||||||
| NC score | 0.020366 (rank : 69) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.020232 (rank : 70) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.019809 (rank : 71) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ANR33_MOUSE
|
||||||
| NC score | 0.019667 (rank : 72) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXP5, Q8BXP4, Q8R1Z1 | Gene names | Ankrd33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.019420 (rank : 73) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
FBW1B_HUMAN
|
||||||
| NC score | 0.018549 (rank : 74) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKB1, Q9P2S8, Q9P2S9, Q9Y4C6 | Gene names | FBXW11, BTRCP2, FBW1B, FBXW1B, KIAA0696 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 11 (F-box/WD repeat protein 1B) (F-box and WD repeats protein beta-TrCP2). | |||||
|
SVIL_HUMAN
|
||||||
| NC score | 0.018416 (rank : 75) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
PAXI_HUMAN
|
||||||
| NC score | 0.018010 (rank : 76) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
PAXI_MOUSE
|
||||||
| NC score | 0.017278 (rank : 77) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.016919 (rank : 78) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
PIAS1_MOUSE
|
||||||
| NC score | 0.016584 (rank : 79) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
CLAT_HUMAN
|
||||||
| NC score | 0.015577 (rank : 80) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28329, Q16488, Q9BQ23, Q9BQ35, Q9BQE1 | Gene names | CHAT | |||
|
Domain Architecture |

|
|||||
| Description | Choline O-acetyltransferase (EC 2.3.1.6) (CHOACTase) (Choline acetylase) (ChAT). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.015358 (rank : 81) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.015004 (rank : 82) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.014938 (rank : 83) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.014689 (rank : 84) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
RAG1_MOUSE
|
||||||
| NC score | 0.014160 (rank : 85) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
FBW1A_HUMAN
|
||||||
| NC score | 0.013772 (rank : 86) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y297, Q9Y213 | Gene names | BTRC, BTRCP, FBW1A, FBXW1A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 1A (F-box and WD repeats protein beta-TrCP) (E3RSIkappaB) (pIkappaBalpha-E3 receptor subunit). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.013645 (rank : 87) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.013170 (rank : 88) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.012877 (rank : 89) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.012626 (rank : 90) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.012596 (rank : 91) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
LRRK1_MOUSE
|
||||||
| NC score | 0.011477 (rank : 92) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
TNR21_HUMAN
|
||||||
| NC score | 0.011200 (rank : 93) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.011026 (rank : 94) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.010792 (rank : 95) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
M3K4_HUMAN
|
||||||
| NC score | 0.010486 (rank : 96) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6R4, Q92612 | Gene names | MAP3K4, KIAA0213, MAPKKK4, MEKK4, MTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4) (MAP three kinase 1). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.010235 (rank : 97) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.009925 (rank : 98) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.009575 (rank : 99) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
KIFC1_HUMAN
|
||||||
| NC score | 0.009495 (rank : 100) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | 0.007700 (rank : 101) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.006633 (rank : 102) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.006625 (rank : 103) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.006266 (rank : 104) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.006006 (rank : 105) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.005568 (rank : 106) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.005198 (rank : 107) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.004831 (rank : 108) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | -0.000717 (rank : 109) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||