Please be patient as the page loads
|
F261_HUMAN
|
||||||
| SwissProt Accessions | P16118, Q99951 | Gene names | PFKFB1, F6PK, PFRX | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (6PF-2-K/Fru- 2,6-P2ASE liver isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
F261_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16118, Q99951 | Gene names | PFKFB1, F6PK, PFRX | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (6PF-2-K/Fru- 2,6-P2ASE liver isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991632 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60825, O60824, Q9H3P1 | Gene names | PFKFB2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F263_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992598 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
F264_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991853 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16877 | Gene names | PFKFB4 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (6PF-2-K/Fru- 2,6-P2ASE testis-type isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_MOUSE
|
||||||
| θ value | 5.00115e-184 (rank : 5) | NC score | 0.992175 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70265, Q8VEI9 | Gene names | Pfkfb2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
RRFM_MOUSE
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.061433 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6S7 | Gene names | Mrrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
ARF6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.038830 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |
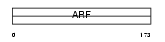
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.038830 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |
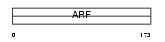
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
PEX5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.027367 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.010133 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
PGAM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.083162 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |
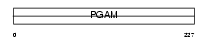
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.083152 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |
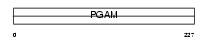
|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.080712 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |
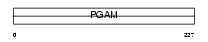
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
RRFM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.055927 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96E11, Q5RKT1, Q5T7T0, Q5T7T1, Q5T7T2, Q5T7T3, Q5T7T4, Q5T7T5 | Gene names | MRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
XAB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.025189 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCN4, O76004 | Gene names | XAB1, MBDIN | |||
|
Domain Architecture |
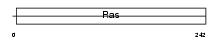
|
|||||
| Description | XPA-binding protein 1. | |||||
|
ARF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.027622 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84077, P10947, P32889 | Gene names | ARF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 1. | |||||
|
ARF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.027622 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84078, P10947, P32889 | Gene names | Arf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 1. | |||||
|
ARF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.027239 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BSL7, P10947, P16500, Q3TJ37, Q91VR9 | Gene names | Arf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 2. | |||||
|
ARF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.027198 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61204, P16587 | Gene names | ARF3 | |||
|
Domain Architecture |
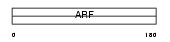
|
|||||
| Description | ADP-ribosylation factor 3. | |||||
|
ARF3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.027198 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61205, P16587 | Gene names | Arf3 | |||
|
Domain Architecture |
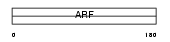
|
|||||
| Description | ADP-ribosylation factor 3. | |||||
|
PGAM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.077733 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |
|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
ARF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.026404 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84085, P26437 | Gene names | ARF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 5. | |||||
|
ARF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.026404 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84084, P26437 | Gene names | Arf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 5. | |||||
|
CN37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.060766 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |
|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
PGAM4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.050681 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
F261_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16118, Q99951 | Gene names | PFKFB1, F6PK, PFRX | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (6PF-2-K/Fru- 2,6-P2ASE liver isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F263_HUMAN
|
||||||
| NC score | 0.992598 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_MOUSE
|
||||||
| NC score | 0.992175 (rank : 3) | θ value | 5.00115e-184 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70265, Q8VEI9 | Gene names | Pfkfb2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F264_HUMAN
|
||||||
| NC score | 0.991853 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16877 | Gene names | PFKFB4 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (6PF-2-K/Fru- 2,6-P2ASE testis-type isozyme) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
F262_HUMAN
|
||||||
| NC score | 0.991632 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60825, O60824, Q9H3P1 | Gene names | PFKFB2 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (6PF-2-K/Fru- 2,6-P2ASE heart-type isozyme) (PFK-2/FBPase-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)]. | |||||
|
PGAM1_HUMAN
|
||||||
| NC score | 0.083162 (rank : 6) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18669, Q9BWC0 | Gene names | PGAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM1_MOUSE
|
||||||
| NC score | 0.083152 (rank : 7) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ1, Q9D6W0, Q9ERT3 | Gene names | Pgam1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 1 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1). | |||||
|
PGAM2_MOUSE
|
||||||
| NC score | 0.080712 (rank : 8) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70250 | Gene names | Pgam2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
PGAM2_HUMAN
|
||||||
| NC score | 0.077733 (rank : 9) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15259 | Gene names | PGAM2, PGAMM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoglycerate mutase 2 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13) (Phosphoglycerate mutase isozyme M) (PGAM-M) (BPG-dependent PGAM 2) (Muscle-specific phosphoglycerate mutase). | |||||
|
RRFM_MOUSE
|
||||||
| NC score | 0.061433 (rank : 10) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6S7 | Gene names | Mrrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
CN37_HUMAN
|
||||||
| NC score | 0.060766 (rank : 11) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09543 | Gene names | CNP | |||
|
Domain Architecture |

|
|||||
| Description | 2',3'-cyclic-nucleotide 3'-phosphodiesterase (EC 3.1.4.37) (CNP) (CNPase). | |||||
|
RRFM_HUMAN
|
||||||
| NC score | 0.055927 (rank : 12) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96E11, Q5RKT1, Q5T7T0, Q5T7T1, Q5T7T2, Q5T7T3, Q5T7T4, Q5T7T5 | Gene names | MRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
PGAM4_HUMAN
|
||||||
| NC score | 0.050681 (rank : 13) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Y7, Q8NI24, Q8NI25, Q8NI26 | Gene names | PGAM4, PGAM3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phosphoglycerate mutase 4 (EC 5.4.2.1) (EC 5.4.2.4) (EC 3.1.3.13). | |||||
|
ARF6_HUMAN
|
||||||
| NC score | 0.038830 (rank : 14) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| NC score | 0.038830 (rank : 15) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF1_HUMAN
|
||||||
| NC score | 0.027622 (rank : 16) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84077, P10947, P32889 | Gene names | ARF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 1. | |||||
|
ARF1_MOUSE
|
||||||
| NC score | 0.027622 (rank : 17) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84078, P10947, P32889 | Gene names | Arf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 1. | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.027367 (rank : 18) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
ARF2_MOUSE
|
||||||
| NC score | 0.027239 (rank : 19) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BSL7, P10947, P16500, Q3TJ37, Q91VR9 | Gene names | Arf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 2. | |||||
|
ARF3_HUMAN
|
||||||
| NC score | 0.027198 (rank : 20) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61204, P16587 | Gene names | ARF3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 3. | |||||
|
ARF3_MOUSE
|
||||||
| NC score | 0.027198 (rank : 21) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61205, P16587 | Gene names | Arf3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 3. | |||||
|
ARF5_HUMAN
|
||||||
| NC score | 0.026404 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84085, P26437 | Gene names | ARF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 5. | |||||
|
ARF5_MOUSE
|
||||||
| NC score | 0.026404 (rank : 23) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P84084, P26437 | Gene names | Arf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor 5. | |||||
|
XAB1_HUMAN
|
||||||
| NC score | 0.025189 (rank : 24) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCN4, O76004 | Gene names | XAB1, MBDIN | |||
|
Domain Architecture |

|
|||||
| Description | XPA-binding protein 1. | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.010133 (rank : 25) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||