Please be patient as the page loads
|
KR134_HUMAN
|
||||||
| SwissProt Accessions | Q3LI77 | Gene names | KRTAP13-4, KAP13.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KR134_HUMAN
|
||||||
| θ value | 1.18549e-84 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q3LI77 | Gene names | KRTAP13-4, KAP13.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-4. | |||||
|
KR131_HUMAN
|
||||||
| θ value | 2.24614e-67 (rank : 2) | NC score | 0.941506 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8IUC0, Q3LI79 | Gene names | KRTAP13-1, KAP13.1, KRTAP13.1 | |||
|
Domain Architecture |
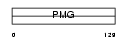
|
|||||
| Description | Keratin-associated protein 13-1 (High sulfur keratin-associated protein 13.1). | |||||
|
KR132_HUMAN
|
||||||
| θ value | 5.37103e-53 (rank : 3) | NC score | 0.958474 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52LG2 | Gene names | KRTAP13-2, KAP13.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-2. | |||||
|
KR133_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 4) | NC score | 0.929777 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3SY46, Q3LI78 | Gene names | KRTAP13-3, KAP13.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-3. | |||||
|
KR151_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 5) | NC score | 0.840822 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3LI76 | Gene names | KRTAP15-1, KAP15.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 15-1. | |||||
|
KR111_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 6) | NC score | 0.780089 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |
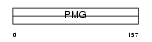
|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
KR412_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 7) | NC score | 0.537685 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 8) | NC score | 0.653682 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 9) | NC score | 0.490513 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 10) | NC score | 0.479771 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 11) | NC score | 0.508653 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 12) | NC score | 0.498769 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 13) | NC score | 0.496705 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 14) | NC score | 0.445995 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.472049 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
KR410_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.605581 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR414_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.533777 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.482155 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KR171_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 19) | NC score | 0.215858 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR106_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 20) | NC score | 0.387902 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR108_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 21) | NC score | 0.392313 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 22) | NC score | 0.479347 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR413_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 23) | NC score | 0.574915 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 24) | NC score | 0.428773 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KR102_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.409282 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 26) | NC score | 0.379742 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 27) | NC score | 0.434508 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 28) | NC score | 0.289693 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR109_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 29) | NC score | 0.411212 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 30) | NC score | 0.275628 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 31) | NC score | 0.216913 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 32) | NC score | 0.240287 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KR122_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 33) | NC score | 0.567181 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 34) | NC score | 0.248124 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KR101_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 35) | NC score | 0.348406 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR261_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 36) | NC score | 0.637816 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
KR10A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 37) | NC score | 0.516700 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 38) | NC score | 0.278898 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA15_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 39) | NC score | 0.470867 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 40) | NC score | 0.217938 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR103_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.526253 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.196197 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.381802 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR212_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.492844 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI59 | Gene names | KRTAP21-2, KAP21.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-2. | |||||
|
KRA13_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.429955 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 46) | NC score | 0.226175 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KR510_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 47) | NC score | 0.264589 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR241_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 48) | NC score | 0.392429 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3LI83 | Gene names | KRTAP24-1, KAP24.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 24-1. | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 49) | NC score | 0.057643 (rank : 130) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 50) | NC score | 0.063789 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
KRA54_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.204905 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 0.125558 (rank : 52) | NC score | 0.568546 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.117528 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.060388 (rank : 128) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
KR123_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.563985 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.046488 (rank : 152) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.124050 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
KR193_HUMAN
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.526309 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
KRHB4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.017451 (rank : 159) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99M73 | Gene names | Krthb4, Krt2-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4) (65 kDa type II keratin). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.043760 (rank : 154) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.044119 (rank : 153) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
KR121_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.323277 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KR195_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.496640 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI72 | Gene names | KRTAP19-5, KAP19.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-5. | |||||
|
GNPTA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.021516 (rank : 157) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.105943 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
KR191_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.409868 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUB9, Q3LI75 | Gene names | KRTAP19-1, KAP19.1, KRTAP19.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-1 (High tyrosine-glycine keratin- associated protein 19.1). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.096400 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.017888 (rank : 158) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.053661 (rank : 144) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
KR211_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.355215 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI58 | Gene names | KRTAP21-1, KAP21.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-1. | |||||
|
TIE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.000895 (rank : 164) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.191699 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.030291 (rank : 156) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
KI18A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.003146 (rank : 163) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
KR124_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.539174 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.062529 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.040831 (rank : 155) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
K1H2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.071211 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |
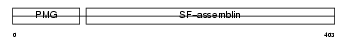
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
K2C4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.011813 (rank : 161) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.015151 (rank : 160) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
TIMD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.009281 (rank : 162) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5QNS5, Q7TPU2, Q8VIM1, Q8VIM2 | Gene names | Havcr1, Tim1, Timd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 homolog precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.089580 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.068011 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.160547 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.055956 (rank : 136) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.054504 (rank : 141) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051334 (rank : 149) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.055426 (rank : 137) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051280 (rank : 150) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052074 (rank : 147) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.056585 (rank : 131) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.058787 (rank : 129) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.064966 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.056165 (rank : 134) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.112322 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.115151 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.055258 (rank : 139) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.078164 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
GRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.136732 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
GRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.130198 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
HHIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.102000 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054454 (rank : 142) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.054874 (rank : 140) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
KR192_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.339714 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LHN2 | Gene names | KRTAP19-2, KAP19.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-2. | |||||
|
KR194_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.183978 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI73 | Gene names | KRTAP19-4, KAP19.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-4. | |||||
|
KR196_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.066450 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI70 | Gene names | KRTAP19-6, KAP19.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-6. | |||||
|
KR197_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.328147 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
KR202_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.287090 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KRA32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.167582 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BYR7 | Gene names | KRTAP3-2, KAP3.2, KRTAP3.2 | |||
|
Domain Architecture |
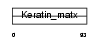
|
|||||
| Description | Keratin-associated protein 3-2 (Keratin-associated protein 3.2) (High sulfur keratin-associated protein 3.2). | |||||
|
KRA33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.172380 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR6, Q52LP0 | Gene names | KRTAP3-3, KAP3.3, KRTAP3.3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 3-3 (Keratin-associated protein 3.3) (High sulfur keratin-associated protein 3.3). | |||||
|
KRA61_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.330639 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI64 | Gene names | KRTAP6-1, KAP6.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-1. | |||||
|
KRA62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.541734 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI66 | Gene names | KRTAP6-2, KAP6.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-2. | |||||
|
KRA63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.176634 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI67 | Gene names | KRTAP6-3, KAP6.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-3. | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052465 (rank : 146) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.090382 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.086884 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051953 (rank : 148) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056060 (rank : 135) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.056308 (rank : 133) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.065347 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.073112 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.062652 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.078718 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053797 (rank : 143) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.065832 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.110667 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NAGPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.084360 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.120981 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.211238 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.055378 (rank : 138) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061418 (rank : 127) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.063667 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053171 (rank : 145) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065301 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.070271 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.064163 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.064660 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.066778 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
RECK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.071569 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
RECK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.069221 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.110990 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.094927 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.148350 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056563 (rank : 132) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.103290 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.101873 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.066185 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.068166 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TENA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.087520 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.079239 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TENR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.062637 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TENR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.068115 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
TENX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.085302 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TRI42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.093681 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
USH2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.061657 (rank : 126) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.066393 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050617 (rank : 151) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
WFDC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.148662 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.218384 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.100965 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.100819 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.062933 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.107127 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.212879 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KR134_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.18549e-84 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q3LI77 | Gene names | KRTAP13-4, KAP13.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-4. | |||||
|
KR132_HUMAN
|
||||||
| NC score | 0.958474 (rank : 2) | θ value | 5.37103e-53 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52LG2 | Gene names | KRTAP13-2, KAP13.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-2. | |||||
|
KR131_HUMAN
|
||||||
| NC score | 0.941506 (rank : 3) | θ value | 2.24614e-67 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8IUC0, Q3LI79 | Gene names | KRTAP13-1, KAP13.1, KRTAP13.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 13-1 (High sulfur keratin-associated protein 13.1). | |||||
|
KR133_HUMAN
|
||||||
| NC score | 0.929777 (rank : 4) | θ value | 4.88048e-38 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3SY46, Q3LI78 | Gene names | KRTAP13-3, KAP13.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 13-3. | |||||
|
KR151_HUMAN
|
||||||
| NC score | 0.840822 (rank : 5) | θ value | 5.60996e-18 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3LI76 | Gene names | KRTAP15-1, KAP15.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 15-1. | |||||
|
KR111_HUMAN
|
||||||
| NC score | 0.780089 (rank : 6) | θ value | 1.29331e-14 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.653682 (rank : 7) | θ value | 1.53129e-07 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KR261_HUMAN
|
||||||
| NC score | 0.637816 (rank : 8) | θ value | 0.00175202 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.605581 (rank : 9) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR413_HUMAN
|
||||||
| NC score | 0.574915 (rank : 10) | θ value | 0.000270298 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.568546 (rank : 11) | θ value | 0.125558 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KR122_HUMAN
|
||||||
| NC score | 0.567181 (rank : 12) | θ value | 0.000786445 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
KR123_HUMAN
|
||||||
| NC score | 0.563985 (rank : 13) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KRA62_HUMAN
|
||||||
| NC score | 0.541734 (rank : 14) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI66 | Gene names | KRTAP6-2, KAP6.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-2. | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.539174 (rank : 15) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.537685 (rank : 16) | θ value | 6.87365e-08 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.533777 (rank : 17) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR193_HUMAN
|
||||||
| NC score | 0.526309 (rank : 18) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4W3 | Gene names | KRTAP19-3, KAP19.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-3 (Glycine/tyrosine-rich protein) (GTHRP). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.526253 (rank : 19) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR10A_HUMAN
|
||||||
| NC score | 0.516700 (rank : 20) | θ value | 0.00228821 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.508653 (rank : 21) | θ value | 2.88788e-06 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.498769 (rank : 22) | θ value | 6.43352e-06 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.496705 (rank : 23) | θ value | 5.44631e-05 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KR195_HUMAN
|
||||||
| NC score | 0.496640 (rank : 24) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3LI72 | Gene names | KRTAP19-5, KAP19.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-5. | |||||
|
KR212_HUMAN
|
||||||
| NC score | 0.492844 (rank : 25) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI59 | Gene names | KRTAP21-2, KAP21.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-2. | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.490513 (rank : 26) | θ value | 1.53129e-07 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.482155 (rank : 27) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.479771 (rank : 28) | θ value | 1.29631e-06 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.479347 (rank : 29) | θ value | 0.000270298 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.472049 (rank : 30) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
KRA15_HUMAN
|
||||||
| NC score | 0.470867 (rank : 31) | θ value | 0.00298849 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.445995 (rank : 32) | θ value | 7.1131e-05 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.434508 (rank : 33) | θ value | 0.000461057 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.429955 (rank : 34) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.428773 (rank : 35) | θ value | 0.000270298 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.411212 (rank : 36) | θ value | 0.000602161 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR191_HUMAN
|
||||||
| NC score | 0.409868 (rank : 37) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUB9, Q3LI75 | Gene names | KRTAP19-1, KAP19.1, KRTAP19.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-1 (High tyrosine-glycine keratin- associated protein 19.1). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.409282 (rank : 38) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR241_HUMAN
|
||||||
| NC score | 0.392429 (rank : 39) | θ value | 0.0252991 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3LI83 | Gene names | KRTAP24-1, KAP24.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 24-1. | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.392313 (rank : 40) | θ value | 0.000270298 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.387902 (rank : 41) | θ value | 0.000270298 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.381802 (rank : 42) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.379742 (rank : 43) | θ value | 0.000461057 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR211_HUMAN
|
||||||
| NC score | 0.355215 (rank : 44) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3LI58 | Gene names | KRTAP21-1, KAP21.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 21-1. | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.348406 (rank : 45) | θ value | 0.00134147 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR192_HUMAN
|
||||||
| NC score | 0.339714 (rank : 46) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LHN2 | Gene names | KRTAP19-2, KAP19.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-2. | |||||
|
KRA61_HUMAN
|
||||||
| NC score | 0.330639 (rank : 47) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI64 | Gene names | KRTAP6-1, KAP6.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-1. | |||||
|
KR197_HUMAN
|
||||||
| NC score | 0.328147 (rank : 48) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3SYF9 | Gene names | KRTAP19-7, KAP19.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-7. | |||||
|
KR121_HUMAN
|
||||||
| NC score | 0.323277 (rank : 49) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.289693 (rank : 50) | θ value | 0.000602161 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR202_HUMAN
|
||||||
| NC score | 0.287090 (rank : 51) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI61 | Gene names | KRTAP20-2, KAP20.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 20-2. | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.278898 (rank : 52) | θ value | 0.00228821 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.275628 (rank : 53) | θ value | 0.000602161 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.264589 (rank : 54) | θ value | 0.0193708 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.248124 (rank : 55) | θ value | 0.00102713 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.240287 (rank : 56) | θ value | 0.000602161 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.226175 (rank : 57) | θ value | 0.0148317 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.218384 (rank : 58) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.217938 (rank : 59) | θ value | 0.00509761 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.216913 (rank : 60) | θ value | 0.000602161 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.215858 (rank : 61) | θ value | 0.00020696 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.212879 (rank : 62) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.211238 (rank : 63) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.204905 (rank : 64) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.196197 (rank : 65) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.191699 (rank : 66) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KR194_HUMAN
|
||||||
| NC score | 0.183978 (rank : 67) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI73 | Gene names | KRTAP19-4, KAP19.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-4. | |||||
|
KRA63_HUMAN
|
||||||
| NC score | 0.176634 (rank : 68) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI67 | Gene names | KRTAP6-3, KAP6.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 6-3. | |||||
|
KRA33_HUMAN
|
||||||
| NC score | 0.172380 (rank : 69) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR6, Q52LP0 | Gene names | KRTAP3-3, KAP3.3, KRTAP3.3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 3-3 (Keratin-associated protein 3.3) (High sulfur keratin-associated protein 3.3). | |||||
|
KRA32_HUMAN
|
||||||
| NC score | 0.167582 (rank : 70) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BYR7 | Gene names | KRTAP3-2, KAP3.2, KRTAP3.2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 3-2 (Keratin-associated protein 3.2) (High sulfur keratin-associated protein 3.2). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.160547 (rank : 71) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
WFDC2_HUMAN
|
||||||
| NC score | 0.148662 (rank : 72) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.148350 (rank : 73) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.136732 (rank : 74) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.130198 (rank : 75) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.124050 (rank : 76) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.120981 (rank : 77) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.117528 (rank : 78) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.115151 (rank : 79) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.112322 (rank : 80) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.110990 (rank : 81) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.110667 (rank : 82) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.107127 (rank : 83) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.105943 (rank : 84) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.103290 (rank : 85) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
HHIP_HUMAN
|
||||||
| NC score | 0.102000 (rank : 86) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.101873 (rank : 87) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.100965 (rank : 88) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.100819 (rank : 89) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.096400 (rank : 90) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.094927 (rank : 91) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
TRI42_MOUSE
|
||||||
| NC score | 0.093681 (rank : 92) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.090382 (rank : 93) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.089580 (rank : 94) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.087520 (rank : 95) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.086884 (rank : 96) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.085302 (rank : 97) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
NAGPA_HUMAN
|
||||||
| NC score | 0.084360 (rank : 98) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.079239 (rank : 99) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.078718 (rank : 100) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.078164 (rank : 101) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.073112 (rank : 102) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.071569 (rank : 103) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
K1H2_HUMAN
|
||||||
| NC score | 0.071211 (rank : 104) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.070271 (rank : 105) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.069221 (rank : 106) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.068166 (rank : 107) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.068115 (rank : 108) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.068011 (rank : 109) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.066778 (rank : 110) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
KR196_HUMAN
|
||||||
| NC score | 0.066450 (rank : 111) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q3LI70 | Gene names | KRTAP19-6, KAP19.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 19-6. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.066393 (rank : 112) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.066185 (rank : 113) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.065832 (rank : 114) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.065347 (rank : 115) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.065301 (rank : 116) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.064966 (rank : 117) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.064660 (rank : 118) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.064163 (rank : 119) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.063789 (rank : 120) | θ value | 0.0330416 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.063667 (rank : 121) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.062933 (rank : 122) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.062652 (rank : 123) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.062637 (rank : 124) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.062529 (rank : 125) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.061657 (rank : 126) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.061418 (rank : 127) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.060388 (rank : 128) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.058787 (rank : 129) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.057643 (rank : 130) | θ value | 0.0330416 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.056585 (rank : 131) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.056563 (rank : 132) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.056308 (rank : 133) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.056165 (rank : 134) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.056060 (rank : 135) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.055956 (rank : 136) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.055426 (rank : 137) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.055378 (rank : 138) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.055258 (rank : 139) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.054874 (rank : 140) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.054504 (rank : 141) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.054454 (rank : 142) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.053797 (rank : 143) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.053661 (rank : 144) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.053171 (rank : 145) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.052465 (rank : 146) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.052074 (rank : 147) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.051953 (rank : 148) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.051334 (rank : 149) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.051280 (rank : 150) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.050617 (rank : 151) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.046488 (rank : 152) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.044119 (rank : 153) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.043760 (rank : 154) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.040831 (rank : 155) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.030291 (rank : 156) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
GNPTA_HUMAN
|
||||||
| NC score | 0.021516 (rank : 157) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.017888 (rank : 158) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
KRHB4_MOUSE
|
||||||
| NC score | 0.017451 (rank : 159) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99M73 | Gene names | Krthb4, Krt2-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb4 (Hair keratin, type II Hb4) (65 kDa type II keratin). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.015151 (rank : 160) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.011813 (rank : 161) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
TIMD1_MOUSE
|
||||||
| NC score | 0.009281 (rank : 162) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5QNS5, Q7TPU2, Q8VIM1, Q8VIM2 | Gene names | Havcr1, Tim1, Timd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 homolog precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1). | |||||
|
KI18A_HUMAN
|
||||||
| NC score | 0.003146 (rank : 163) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
TIE2_MOUSE
|
||||||
| NC score | 0.000895 (rank : 164) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||