Please be patient as the page loads
|
DDHD1_HUMAN
|
||||||
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDHD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993365 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 3) | NC score | 0.602223 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 4) | NC score | 0.560949 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 5) | NC score | 0.359202 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 6) | NC score | 0.361828 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 7) | NC score | 0.337168 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM3_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 8) | NC score | 0.379765 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 9) | NC score | 0.376992 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 10) | NC score | 0.333843 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.083934 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.051095 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
T240L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.048434 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
PHOS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.050458 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.002265 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.016488 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.019565 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
K1C12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.008404 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.023008 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.016737 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IQEC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.013167 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.016198 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.002236 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.022996 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.022255 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.024751 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.018310 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
CACB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.012053 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
DLGP3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.021277 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
AL7A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.008361 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.024472 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
E2AK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.002020 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
MCR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.008564 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
MCR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.008830 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
PRP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.022451 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P2Q9, O14547, O75965 | Gene names | PRPF8, PRPC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8) (PRP8 homolog) (220 kDa U5 snRNP-specific protein) (p220). | |||||
|
PRP8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.022452 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PV0, Q5ND30, Q5ND31, Q7TQK2, Q8BRZ5, Q8K001 | Gene names | Prpf8, Prp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.016667 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.015740 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.005881 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.014528 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
PIPNA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.130726 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.131267 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.129146 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |
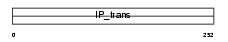
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.129537 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |
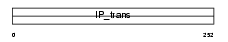
|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_MOUSE
|
||||||
| NC score | 0.993365 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.602223 (rank : 3) | θ value | 8.08199e-25 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.560949 (rank : 4) | θ value | 7.56453e-23 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
PITM3_MOUSE
|
||||||
| NC score | 0.379765 (rank : 5) | θ value | 1.29631e-06 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UHE1, Q3UH22, Q5RIT9 | Gene names | Pitpnm3, Nir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM3_HUMAN
|
||||||
| NC score | 0.376992 (rank : 6) | θ value | 1.69304e-06 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ71, Q59GH9, Q9NPQ4 | Gene names | PITPNM3, NIR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 3 (Phosphatidylinositol transfer protein, membrane-associated 3) (Pyk2 N-terminal domain-interacting receptor 1) (NIR-1). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.361828 (rank : 7) | θ value | 4.0297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.359202 (rank : 8) | θ value | 4.0297e-08 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
PITM2_MOUSE
|
||||||
| NC score | 0.337168 (rank : 9) | θ value | 1.29631e-06 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZPQ6, Q9R1P5 | Gene names | Pitpnm2, Kiaa1457, Nir3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3) (Drosophila retinal degeneration B homolog 2) (RdgB2). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.333843 (rank : 10) | θ value | 2.88788e-06 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
PIPNA_MOUSE
|
||||||
| NC score | 0.131267 (rank : 11) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53810 | Gene names | Pitpna, Pitpn | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNA_HUMAN
|
||||||
| NC score | 0.130726 (rank : 12) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00169 | Gene names | PITPNA, PITPN | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein alpha isoform (PtdIns transfer protein alpha) (PtdInsTP) (PI-TP-alpha). | |||||
|
PIPNB_MOUSE
|
||||||
| NC score | 0.129537 (rank : 13) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53811 | Gene names | Pitpnb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
PIPNB_HUMAN
|
||||||
| NC score | 0.129146 (rank : 14) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48739, Q8N5W1 | Gene names | PITPNB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol transfer protein beta isoform (PtdIns transfer protein beta) (PtdInsTP) (PI-TP-beta). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.083934 (rank : 15) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.051095 (rank : 16) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
PHOS_MOUSE
|
||||||
| NC score | 0.050458 (rank : 17) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.048434 (rank : 18) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.024751 (rank : 19) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.024472 (rank : 20) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.023008 (rank : 21) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.022996 (rank : 22) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PRP8_MOUSE
|
||||||
| NC score | 0.022452 (rank : 23) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PV0, Q5ND30, Q5ND31, Q7TQK2, Q8BRZ5, Q8K001 | Gene names | Prpf8, Prp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8). | |||||
|
PRP8_HUMAN
|
||||||
| NC score | 0.022451 (rank : 24) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P2Q9, O14547, O75965 | Gene names | PRPF8, PRPC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-processing-splicing factor 8 (Splicing factor Prp8) (PRP8 homolog) (220 kDa U5 snRNP-specific protein) (p220). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.022255 (rank : 25) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP3_MOUSE
|
||||||
| NC score | 0.021277 (rank : 26) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.019565 (rank : 27) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.018310 (rank : 28) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.016737 (rank : 29) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.016667 (rank : 30) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.016488 (rank : 31) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.016198 (rank : 32) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.015740 (rank : 33) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.014528 (rank : 34) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
IQEC1_HUMAN
|
||||||
| NC score | 0.013167 (rank : 35) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
CACB2_HUMAN
|
||||||
| NC score | 0.012053 (rank : 36) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08289, O00304, Q8WX81, Q96NZ3, Q96NZ4, Q96NZ5, Q9BWU2, Q9HD32, Q9Y340, Q9Y341 | Gene names | CACNB2, CACNLB2, MYSB | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2) (Lambert-Eaton myasthenic syndrome antigen B) (MYSB). | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.008830 (rank : 37) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
MCR_HUMAN
|
||||||
| NC score | 0.008564 (rank : 38) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
K1C12_HUMAN
|
||||||
| NC score | 0.008404 (rank : 39) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
AL7A1_HUMAN
|
||||||
| NC score | 0.008361 (rank : 40) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.005881 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.002265 (rank : 42) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
SP1_HUMAN
|
||||||
| NC score | 0.002236 (rank : 43) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
E2AK4_HUMAN
|
||||||
| NC score | 0.002020 (rank : 44) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||