Please be patient as the page loads
|
PHOS_MOUSE
|
||||||
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PHOS_MOUSE
|
||||||
| θ value | 8.35901e-139 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
PHOS_HUMAN
|
||||||
| θ value | 1.99884e-124 (rank : 2) | NC score | 0.988171 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
PHLP_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 3) | NC score | 0.943764 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PHLP_MOUSE
|
||||||
| θ value | 1.78386e-56 (rank : 4) | NC score | 0.942805 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |
|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PDCL2_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 5) | NC score | 0.603161 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL3_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 6) | NC score | 0.605492 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
PDCL3_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 7) | NC score | 0.590734 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2J4 | Gene names | PDCL3, VIAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1) (HTPHLP). | |||||
|
PDCL2_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 8) | NC score | 0.589145 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
TXND9_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.271796 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQ79, Q3TKD2 | Gene names | Txndc9, Apacd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 9 (ATP-binding protein associated with cell differentiation). | |||||
|
TXND9_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.269814 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14530, Q53RV8, Q6NSF5, Q9BRU6 | Gene names | TXNDC9, APACD | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 9 (Protein 1-4) (ATP-binding protein associated with cell differentiation). | |||||
|
ACON_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.046013 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KI0 | Gene names | Aco2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase). | |||||
|
DDHD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.053677 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.050458 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.047663 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.048234 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
THIOM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.046349 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
ZN213_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.000820 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14771, Q96IS1 | Gene names | ZNF213 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 213 (Putative transcription factor CR53). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.006453 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
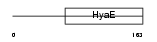
THIOM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.045069 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
ZBED4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.038411 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
ACON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.037190 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99798, O75809 | Gene names | ACO2 | |||
|
Domain Architecture |

|
|||||
| Description | Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase). | |||||
|
DGKI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.005185 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75912, Q9NZ49 | Gene names | DGKI | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase iota (EC 2.7.1.107) (Diglyceride kinase iota) (DGK-iota) (DAG kinase iota). | |||||
|
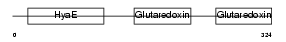
TXNL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.106407 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
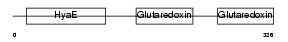
TXNL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.127336 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
PHOS_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.35901e-139 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
PHOS_HUMAN
|
||||||
| NC score | 0.988171 (rank : 2) | θ value | 1.99884e-124 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
PHLP_HUMAN
|
||||||
| NC score | 0.943764 (rank : 3) | θ value | 2.94036e-59 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PHLP_MOUSE
|
||||||
| NC score | 0.942805 (rank : 4) | θ value | 1.78386e-56 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PDCL3_MOUSE
|
||||||
| NC score | 0.605492 (rank : 5) | θ value | 5.62301e-10 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
PDCL2_MOUSE
|
||||||
| NC score | 0.603161 (rank : 6) | θ value | 5.62301e-10 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL3_HUMAN
|
||||||
| NC score | 0.590734 (rank : 7) | θ value | 3.64472e-09 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2J4 | Gene names | PDCL3, VIAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1) (HTPHLP). | |||||
|
PDCL2_HUMAN
|
||||||
| NC score | 0.589145 (rank : 8) | θ value | 6.21693e-09 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
TXND9_MOUSE
|
||||||
| NC score | 0.271796 (rank : 9) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQ79, Q3TKD2 | Gene names | Txndc9, Apacd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 9 (ATP-binding protein associated with cell differentiation). | |||||
|
TXND9_HUMAN
|
||||||
| NC score | 0.269814 (rank : 10) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14530, Q53RV8, Q6NSF5, Q9BRU6 | Gene names | TXNDC9, APACD | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 9 (Protein 1-4) (ATP-binding protein associated with cell differentiation). | |||||
|
TXNL2_MOUSE
|
||||||
| NC score | 0.127336 (rank : 11) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
TXNL2_HUMAN
|
||||||
| NC score | 0.106407 (rank : 12) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
DDHD1_MOUSE
|
||||||
| NC score | 0.053677 (rank : 13) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA3, Q8VEF2 | Gene names | Ddhd1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.050458 (rank : 14) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.048234 (rank : 15) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.047663 (rank : 16) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
THIOM_HUMAN
|
||||||
| NC score | 0.046349 (rank : 17) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
ACON_MOUSE
|
||||||
| NC score | 0.046013 (rank : 18) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KI0 | Gene names | Aco2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase). | |||||
|
THIOM_MOUSE
|
||||||
| NC score | 0.045069 (rank : 19) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
ZBED4_HUMAN
|
||||||
| NC score | 0.038411 (rank : 20) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
ACON_HUMAN
|
||||||
| NC score | 0.037190 (rank : 21) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99798, O75809 | Gene names | ACO2 | |||
|
Domain Architecture |

|
|||||
| Description | Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Citrate hydro-lyase) (Aconitase). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006453 (rank : 22) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
DGKI_HUMAN
|
||||||
| NC score | 0.005185 (rank : 23) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75912, Q9NZ49 | Gene names | DGKI | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase iota (EC 2.7.1.107) (Diglyceride kinase iota) (DGK-iota) (DAG kinase iota). | |||||
|
ZN213_HUMAN
|
||||||
| NC score | 0.000820 (rank : 24) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14771, Q96IS1 | Gene names | ZNF213 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 213 (Putative transcription factor CR53). | |||||