Please be patient as the page loads
|
CSAD_MOUSE
|
||||||
| SwissProt Accessions | Q9DBE0 | Gene names | Csad | |||
|
Domain Architecture |
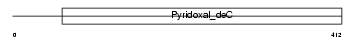
|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CSAD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998994 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y600, Q9UNJ5, Q9Y601 | Gene names | CSAD, CSD | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
CSAD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBE0 | Gene names | Csad | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
DCE1_HUMAN
|
||||||
| θ value | 5.22361e-149 (rank : 3) | NC score | 0.984538 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99259, Q9BU91, Q9UHH4 | Gene names | GAD1, GAD, GAD67 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 1 (EC 4.1.1.15) (Glutamate decarboxylase 67 kDa isoform) (GAD-67) (67 kDa glutamic acid decarboxylase). | |||||
|
DCE2_HUMAN
|
||||||
| θ value | 5.77539e-148 (rank : 4) | NC score | 0.984530 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05329, Q9UD87 | Gene names | GAD2, GAD65 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 2 (EC 4.1.1.15) (Glutamate decarboxylase 65 kDa isoform) (GAD-65) (65 kDa glutamic acid decarboxylase). | |||||
|
DCE1_MOUSE
|
||||||
| θ value | 1.28662e-147 (rank : 5) | NC score | 0.984351 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48318, O08685 | Gene names | Gad1, Gad67 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 1 (EC 4.1.1.15) (Glutamate decarboxylase 67 kDa isoform) (GAD-67) (67 kDa glutamic acid decarboxylase). | |||||
|
DCE2_MOUSE
|
||||||
| θ value | 3.7435e-147 (rank : 6) | NC score | 0.984237 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48320, O35519 | Gene names | Gad2, Gad65 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 2 (EC 4.1.1.15) (Glutamate decarboxylase 65 kDa isoform) (GAD-65) (65 kDa glutamic acid decarboxylase). | |||||
|
DDC_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 7) | NC score | 0.762456 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20711, Q16723 | Gene names | DDC, AADC | |||
|
Domain Architecture |

|
|||||
| Description | Aromatic-L-amino-acid decarboxylase (EC 4.1.1.28) (AADC) (DOPA decarboxylase) (DDC). | |||||
|
DDC_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 8) | NC score | 0.759678 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88533 | Gene names | Ddc | |||
|
Domain Architecture |

|
|||||
| Description | Aromatic-L-amino-acid decarboxylase (EC 4.1.1.28) (AADC) (DOPA decarboxylase) (DDC). | |||||
|
DCHS_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 9) | NC score | 0.725506 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23738 | Gene names | Hdc | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
DCHS_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 10) | NC score | 0.727419 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19113 | Gene names | HDC | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
NFS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.042824 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1J3 | Gene names | Nfs1, Nifs | |||
|
Domain Architecture |
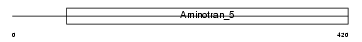
|
|||||
| Description | Cysteine desulfurase, mitochondrial precursor (EC 2.8.1.7) (m-Nfs1). | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.008659 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
CT012_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.016262 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
NFS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.029156 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y697, Q6P0L8, Q9NTZ5, Q9Y481 | Gene names | NFS1, NIFS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine desulfurase, mitochondrial precursor (EC 2.8.1.7). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.008412 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.008405 (rank : 18) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
MSH5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.006722 (rank : 19) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
SGPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.105440 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95470, Q7Z732, Q9ULG8, Q9UN89 | Gene names | SGPL1, KIAA1252 | |||
|
Domain Architecture |
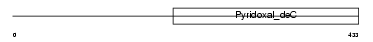
|
|||||
| Description | Sphingosine-1-phosphate lyase 1 (EC 4.1.2.27) (SP-lyase) (hSPL) (Sphingosine-1-phosphate aldolase). | |||||
|
SGPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.082141 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0X7, O54955, Q8C942 | Gene names | Sgpl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine-1-phosphate lyase 1 (EC 4.1.2.27) (SP-lyase) (mSPL) (Sphingosine-1-phosphate aldolase). | |||||
|
CSAD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBE0 | Gene names | Csad | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
CSAD_HUMAN
|
||||||
| NC score | 0.998994 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y600, Q9UNJ5, Q9Y601 | Gene names | CSAD, CSD | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine sulfinic acid decarboxylase (EC 4.1.1.29) (Sulfinoalanine decarboxylase) (Cysteine-sulfinate decarboxylase). | |||||
|
DCE1_HUMAN
|
||||||
| NC score | 0.984538 (rank : 3) | θ value | 5.22361e-149 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99259, Q9BU91, Q9UHH4 | Gene names | GAD1, GAD, GAD67 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 1 (EC 4.1.1.15) (Glutamate decarboxylase 67 kDa isoform) (GAD-67) (67 kDa glutamic acid decarboxylase). | |||||
|
DCE2_HUMAN
|
||||||
| NC score | 0.984530 (rank : 4) | θ value | 5.77539e-148 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05329, Q9UD87 | Gene names | GAD2, GAD65 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 2 (EC 4.1.1.15) (Glutamate decarboxylase 65 kDa isoform) (GAD-65) (65 kDa glutamic acid decarboxylase). | |||||
|
DCE1_MOUSE
|
||||||
| NC score | 0.984351 (rank : 5) | θ value | 1.28662e-147 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48318, O08685 | Gene names | Gad1, Gad67 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 1 (EC 4.1.1.15) (Glutamate decarboxylase 67 kDa isoform) (GAD-67) (67 kDa glutamic acid decarboxylase). | |||||
|
DCE2_MOUSE
|
||||||
| NC score | 0.984237 (rank : 6) | θ value | 3.7435e-147 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48320, O35519 | Gene names | Gad2, Gad65 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate decarboxylase 2 (EC 4.1.1.15) (Glutamate decarboxylase 65 kDa isoform) (GAD-65) (65 kDa glutamic acid decarboxylase). | |||||
|
DDC_HUMAN
|
||||||
| NC score | 0.762456 (rank : 7) | θ value | 1.92365e-26 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20711, Q16723 | Gene names | DDC, AADC | |||
|
Domain Architecture |

|
|||||
| Description | Aromatic-L-amino-acid decarboxylase (EC 4.1.1.28) (AADC) (DOPA decarboxylase) (DDC). | |||||
|
DDC_MOUSE
|
||||||
| NC score | 0.759678 (rank : 8) | θ value | 6.18819e-25 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88533 | Gene names | Ddc | |||
|
Domain Architecture |

|
|||||
| Description | Aromatic-L-amino-acid decarboxylase (EC 4.1.1.28) (AADC) (DOPA decarboxylase) (DDC). | |||||
|
DCHS_HUMAN
|
||||||
| NC score | 0.727419 (rank : 9) | θ value | 3.39556e-23 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19113 | Gene names | HDC | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
DCHS_MOUSE
|
||||||
| NC score | 0.725506 (rank : 10) | θ value | 5.23862e-24 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23738 | Gene names | Hdc | |||
|
Domain Architecture |

|
|||||
| Description | Histidine decarboxylase (EC 4.1.1.22) (HDC). | |||||
|
SGPL1_HUMAN
|
||||||
| NC score | 0.105440 (rank : 11) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95470, Q7Z732, Q9ULG8, Q9UN89 | Gene names | SGPL1, KIAA1252 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine-1-phosphate lyase 1 (EC 4.1.2.27) (SP-lyase) (hSPL) (Sphingosine-1-phosphate aldolase). | |||||
|
SGPL1_MOUSE
|
||||||
| NC score | 0.082141 (rank : 12) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R0X7, O54955, Q8C942 | Gene names | Sgpl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine-1-phosphate lyase 1 (EC 4.1.2.27) (SP-lyase) (mSPL) (Sphingosine-1-phosphate aldolase). | |||||
|
NFS1_MOUSE
|
||||||
| NC score | 0.042824 (rank : 13) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1J3 | Gene names | Nfs1, Nifs | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine desulfurase, mitochondrial precursor (EC 2.8.1.7) (m-Nfs1). | |||||
|
NFS1_HUMAN
|
||||||
| NC score | 0.029156 (rank : 14) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y697, Q6P0L8, Q9NTZ5, Q9Y481 | Gene names | NFS1, NIFS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine desulfurase, mitochondrial precursor (EC 2.8.1.7). | |||||
|
CT012_HUMAN
|
||||||
| NC score | 0.016262 (rank : 15) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVP4, Q4F7X1, Q5QPD9, Q5QPE0, Q68DN8, Q6ZMX9, Q96NF0, Q9H1E0, Q9H442 | Gene names | C20orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf12. | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.008659 (rank : 16) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.008412 (rank : 17) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.008405 (rank : 18) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.006722 (rank : 19) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||