Please be patient as the page loads
|
UBR1_HUMAN
|
||||||
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UBR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997177 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70481, Q5BKR8, Q792M3, Q8BN40, Q8C5K3 | Gene names | Ubr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UBR2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979819 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
UBR2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.980492 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
JAK2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.016270 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60674, O14636, O75297 | Gene names | JAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK2 (EC 2.7.10.2) (Janus kinase 2) (JAK-2). | |||||
|
JAK2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.015067 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62120, Q62124 | Gene names | Jak2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK2 (EC 2.7.10.2) (Janus kinase 2) (JAK-2). | |||||
|
ZN583_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.006287 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.036830 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.036844 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.004067 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
ZN578_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.006223 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96N58 | Gene names | ZNF578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 578. | |||||
|
ZN583_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.005266 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.025752 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CE110_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.026186 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CNN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.016608 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
PP1R8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.039761 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12972, Q5TIF2, Q6PKF6, Q9UBH1, Q9UBZ0 | Gene names | PPP1R8, ARD1, NIPP1 | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear inhibitor of protein phosphatase 1 (NIPP-1) (Protein phosphatase 1 regulatory inhibitor subunit 8) [Includes: Activator of RNA decay (EC 3.1.4.-) (ARD-1)]. | |||||
|
PP1R8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.039737 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3G1, Q8C087 | Gene names | Ppp1r8, Nipp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear inhibitor of protein phosphatase 1 (NIPP-1) (Protein phosphatase 1 regulatory inhibitor subunit 8). | |||||
|
PUR6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.028972 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22234 | Gene names | PAICS, ADE2, AIRC, PAIS | |||
|
Domain Architecture |

|
|||||
| Description | Multifunctional protein ADE2 [Includes: Phosphoribosylaminoimidazole- succinocarboxamide synthase (EC 6.3.2.6) (SAICAR synthetase); Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) (AIR carboxylase) (AIRC)]. | |||||
|
PUR6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.028902 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCL9 | Gene names | Paics | |||
|
Domain Architecture |

|
|||||
| Description | Multifunctional protein ADE2 [Includes: Phosphoribosylaminoimidazole- succinocarboxamide synthase (EC 6.3.2.6) (SAICAR synthetase); Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) (AIR carboxylase) (AIRC)]. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.006315 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN283_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.005954 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N7M2 | Gene names | ZNF283 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 283. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.005560 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.005482 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
EDG4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.010794 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL06 | Gene names | Edg4, Lpa2 | |||
|
Domain Architecture |
|
|||||
| Description | Lysophosphatidic acid receptor Edg-4 (LPA receptor 2) (LPA-2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.006859 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.006294 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
CNN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.014337 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
COPA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.011391 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53621 | Gene names | COPA | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit alpha (Alpha-coat protein) (Alpha-COP) (HEPCOP) (HEP- COP) [Contains: Xenin (Xenopsin-related peptide); Proxenin]. | |||||
|
DYDC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.035468 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.014845 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.012218 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.013754 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
ZN471_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.003850 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX82, O75260, Q8N3V1, Q9P2F1 | Gene names | ZNF471, ERP1, KIAA1396 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 471 (EZFIT-related protein 1). | |||||
|
ZN572_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.004755 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||
|
ZNF34_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.003757 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZ26 | Gene names | ZNF34, KOX32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 34 (Zinc finger protein KOX32). | |||||
|
ENKUR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.014590 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
CNN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.013216 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CP3AG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.002161 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64481 | Gene names | Cyp3a16, Cyp3a-16 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 3A16 (EC 1.14.14.1) (CYPIIIA16). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.003811 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
GNPAT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.009355 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.001793 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
HKR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.004067 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10072, Q9BSW9, Q9UM09 | Gene names | HKR1 | |||
|
Domain Architecture |
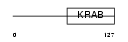
|
|||||
| Description | Krueppel-related zinc finger protein 1 (Protein HKR1). | |||||
|
IFNK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.014244 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W0, Q5T166 | Gene names | IFNK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon kappa precursor (IFN-kappa). | |||||
|
UBR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UBR1_MOUSE
|
||||||
| NC score | 0.997177 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O70481, Q5BKR8, Q792M3, Q8BN40, Q8C5K3 | Gene names | Ubr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UBR2_MOUSE
|
||||||
| NC score | 0.980492 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
UBR2_HUMAN
|
||||||
| NC score | 0.979819 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
PP1R8_HUMAN
|
||||||
| NC score | 0.039761 (rank : 5) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12972, Q5TIF2, Q6PKF6, Q9UBH1, Q9UBZ0 | Gene names | PPP1R8, ARD1, NIPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear inhibitor of protein phosphatase 1 (NIPP-1) (Protein phosphatase 1 regulatory inhibitor subunit 8) [Includes: Activator of RNA decay (EC 3.1.4.-) (ARD-1)]. | |||||
|
PP1R8_MOUSE
|
||||||
| NC score | 0.039737 (rank : 6) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3G1, Q8C087 | Gene names | Ppp1r8, Nipp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear inhibitor of protein phosphatase 1 (NIPP-1) (Protein phosphatase 1 regulatory inhibitor subunit 8). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.036844 (rank : 7) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.036830 (rank : 8) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
DYDC2_HUMAN
|
||||||
| NC score | 0.035468 (rank : 9) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
PUR6_HUMAN
|
||||||
| NC score | 0.028972 (rank : 10) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22234 | Gene names | PAICS, ADE2, AIRC, PAIS | |||
|
Domain Architecture |

|
|||||
| Description | Multifunctional protein ADE2 [Includes: Phosphoribosylaminoimidazole- succinocarboxamide synthase (EC 6.3.2.6) (SAICAR synthetase); Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) (AIR carboxylase) (AIRC)]. | |||||
|
PUR6_MOUSE
|
||||||
| NC score | 0.028902 (rank : 11) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCL9 | Gene names | Paics | |||
|
Domain Architecture |

|
|||||
| Description | Multifunctional protein ADE2 [Includes: Phosphoribosylaminoimidazole- succinocarboxamide synthase (EC 6.3.2.6) (SAICAR synthetase); Phosphoribosylaminoimidazole carboxylase (EC 4.1.1.21) (AIR carboxylase) (AIRC)]. | |||||
|
CE110_MOUSE
|
||||||
| NC score | 0.026186 (rank : 12) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.025752 (rank : 13) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CNN1_HUMAN
|
||||||
| NC score | 0.016608 (rank : 14) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
JAK2_HUMAN
|
||||||
| NC score | 0.016270 (rank : 15) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60674, O14636, O75297 | Gene names | JAK2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK2 (EC 2.7.10.2) (Janus kinase 2) (JAK-2). | |||||
|
JAK2_MOUSE
|
||||||
| NC score | 0.015067 (rank : 16) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62120, Q62124 | Gene names | Jak2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK2 (EC 2.7.10.2) (Janus kinase 2) (JAK-2). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.014845 (rank : 17) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
ENKUR_HUMAN
|
||||||
| NC score | 0.014590 (rank : 18) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
CNN3_MOUSE
|
||||||
| NC score | 0.014337 (rank : 19) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
IFNK_HUMAN
|
||||||
| NC score | 0.014244 (rank : 20) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W0, Q5T166 | Gene names | IFNK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon kappa precursor (IFN-kappa). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.013754 (rank : 21) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CNN1_MOUSE
|
||||||
| NC score | 0.013216 (rank : 22) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.012218 (rank : 23) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
COPA_HUMAN
|
||||||
| NC score | 0.011391 (rank : 24) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53621 | Gene names | COPA | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit alpha (Alpha-coat protein) (Alpha-COP) (HEPCOP) (HEP- COP) [Contains: Xenin (Xenopsin-related peptide); Proxenin]. | |||||
|
EDG4_MOUSE
|
||||||
| NC score | 0.010794 (rank : 25) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL06 | Gene names | Edg4, Lpa2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysophosphatidic acid receptor Edg-4 (LPA receptor 2) (LPA-2). | |||||
|
GNPAT_HUMAN
|
||||||
| NC score | 0.009355 (rank : 26) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.006859 (rank : 27) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.006315 (rank : 28) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.006294 (rank : 29) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ZN583_MOUSE
|
||||||
| NC score | 0.006287 (rank : 30) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||
|
ZN578_HUMAN
|
||||||
| NC score | 0.006223 (rank : 31) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96N58 | Gene names | ZNF578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 578. | |||||
|
ZN283_HUMAN
|
||||||
| NC score | 0.005954 (rank : 32) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N7M2 | Gene names | ZNF283 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 283. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.005560 (rank : 33) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.005482 (rank : 34) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ZN583_HUMAN
|
||||||
| NC score | 0.005266 (rank : 35) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||
|
ZN572_HUMAN
|
||||||
| NC score | 0.004755 (rank : 36) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3I7, Q8N1Q0 | Gene names | ZNF572 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 572. | |||||
|
HKR1_HUMAN
|
||||||
| NC score | 0.004067 (rank : 37) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10072, Q9BSW9, Q9UM09 | Gene names | HKR1 | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-related zinc finger protein 1 (Protein HKR1). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.004067 (rank : 38) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
ZN471_HUMAN
|
||||||
| NC score | 0.003850 (rank : 39) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BX82, O75260, Q8N3V1, Q9P2F1 | Gene names | ZNF471, ERP1, KIAA1396 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 471 (EZFIT-related protein 1). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | 0.003811 (rank : 40) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
ZNF34_HUMAN
|
||||||
| NC score | 0.003757 (rank : 41) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZ26 | Gene names | ZNF34, KOX32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 34 (Zinc finger protein KOX32). | |||||
|
CP3AG_MOUSE
|
||||||
| NC score | 0.002161 (rank : 42) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64481 | Gene names | Cyp3a16, Cyp3a-16 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 3A16 (EC 1.14.14.1) (CYPIIIA16). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.001793 (rank : 43) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||