Please be patient as the page loads
|
SPRE3_HUMAN
|
||||||
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPRE3_MOUSE
|
||||||
| θ value | 5.07123e-136 (rank : 1) | NC score | 0.984363 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE3_HUMAN
|
||||||
| θ value | 9.58617e-127 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE2_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 3) | NC score | 0.938231 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE2_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 4) | NC score | 0.936620 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE1_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 5) | NC score | 0.869022 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 6) | NC score | 0.869107 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPY3_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 7) | NC score | 0.577843 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 8) | NC score | 0.400625 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 9) | NC score | 0.512948 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 10) | NC score | 0.381146 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 11) | NC score | 0.534470 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 12) | NC score | 0.534382 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 13) | NC score | 0.506093 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
SPY1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 14) | NC score | 0.516723 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY4_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 15) | NC score | 0.511676 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |
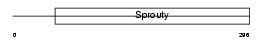
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 16) | NC score | 0.510764 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |
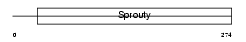
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 17) | NC score | 0.502158 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.484032 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SPY1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.458393 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |
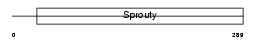
|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.040789 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.014102 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
EPHA1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.009527 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.043396 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.048241 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
FOXL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.017825 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |
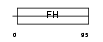
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.042765 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.030556 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
UB7I4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.044028 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.040987 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.036868 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.017699 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.088354 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.087728 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.107688 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.114864 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.035002 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.036123 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
UB7I4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.038013 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |
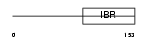
|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.012235 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
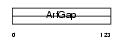
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.010489 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.046799 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.022669 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
YPEL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.020131 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NS1, Q65Z98 | Gene names | YPEL4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein yippee-like 4. | |||||
|
NIPA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.029889 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86WB0, Q75MF3, Q75MF4, Q8N330, Q96F75, Q9HA34, Q9NVX4, Q9P0R0 | Gene names | ZC3HC1, NIPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (hNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.051265 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.009124 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CNTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.007241 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
DOK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.025948 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |
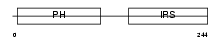
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
ANKK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.002414 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.038905 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CU124_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.024123 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HW9 | Gene names | C21orf124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf124. | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.013721 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.010274 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MADCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.015323 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61826, O35530, Q61278, Q64275, Q8R1M6 | Gene names | Madcam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (mMAdCAM-1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.029209 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.028581 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.019651 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
BEGIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.009043 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.012076 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.006751 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
HXD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.000963 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.014403 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.041939 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.002392 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.006040 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
HOME3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.066278 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC5, O14580, O95350, Q9NSB9, Q9NSC0 | Gene names | HOMER3 | |||
|
Domain Architecture |
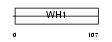
|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
HOME3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.067940 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JP6, Q9Z215 | Gene names | Homer3 | |||
|
Domain Architecture |
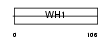
|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
PTTG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.064146 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R143, Q3TVT1, Q8BJ96, Q8N7P0 | Gene names | Pttg1ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pituitary tumor-transforming gene 1 protein-interacting protein precursor (Pituitary tumor-transforming gene protein-binding factor) (PTTG-binding factor) (PBF). | |||||
|
SPRE3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.58617e-127 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.984363 (rank : 2) | θ value | 5.07123e-136 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE2_MOUSE
|
||||||
| NC score | 0.938231 (rank : 3) | θ value | 4.22625e-74 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE2_HUMAN
|
||||||
| NC score | 0.936620 (rank : 4) | θ value | 7.97034e-73 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE1_HUMAN
|
||||||
| NC score | 0.869107 (rank : 5) | θ value | 6.61148e-27 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_MOUSE
|
||||||
| NC score | 0.869022 (rank : 6) | θ value | 1.02001e-27 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPY3_HUMAN
|
||||||
| NC score | 0.577843 (rank : 7) | θ value | 5.08577e-11 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.534470 (rank : 8) | θ value | 1.25267e-09 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.534382 (rank : 9) | θ value | 1.25267e-09 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY1_HUMAN
|
||||||
| NC score | 0.516723 (rank : 10) | θ value | 8.11959e-09 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.512948 (rank : 11) | θ value | 7.34386e-10 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
SPY4_HUMAN
|
||||||
| NC score | 0.511676 (rank : 12) | θ value | 2.61198e-07 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_MOUSE
|
||||||
| NC score | 0.510764 (rank : 13) | θ value | 2.61198e-07 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.506093 (rank : 14) | θ value | 3.64472e-09 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.502158 (rank : 15) | θ value | 1.29631e-06 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.484032 (rank : 16) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SPY1_MOUSE
|
||||||
| NC score | 0.458393 (rank : 17) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.400625 (rank : 18) | θ value | 3.29651e-10 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.381146 (rank : 19) | θ value | 9.59137e-10 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.114864 (rank : 20) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.107688 (rank : 21) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.088354 (rank : 22) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.087728 (rank : 23) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
HOME3_MOUSE
|
||||||
| NC score | 0.067940 (rank : 24) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JP6, Q9Z215 | Gene names | Homer3 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
HOME3_HUMAN
|
||||||
| NC score | 0.066278 (rank : 25) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSC5, O14580, O95350, Q9NSB9, Q9NSC0 | Gene names | HOMER3 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
PTTG_MOUSE
|
||||||
| NC score | 0.064146 (rank : 26) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R143, Q3TVT1, Q8BJ96, Q8N7P0 | Gene names | Pttg1ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pituitary tumor-transforming gene 1 protein-interacting protein precursor (Pituitary tumor-transforming gene protein-binding factor) (PTTG-binding factor) (PBF). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.051265 (rank : 27) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.048241 (rank : 28) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.046799 (rank : 29) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
UB7I4_HUMAN
|
||||||
| NC score | 0.044028 (rank : 30) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.043396 (rank : 31) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.042765 (rank : 32) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.041939 (rank : 33) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.040987 (rank : 34) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.040789 (rank : 35) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.038905 (rank : 36) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
UB7I4_MOUSE
|
||||||
| NC score | 0.038013 (rank : 37) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925F3, Q6A0B4 | Gene names | Rnf144, Kiaa0161, Ubce7ip4, Uip4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.036868 (rank : 38) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036123 (rank : 39) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.035002 (rank : 40) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.030556 (rank : 41) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
NIPA_HUMAN
|
||||||
| NC score | 0.029889 (rank : 42) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86WB0, Q75MF3, Q75MF4, Q8N330, Q96F75, Q9HA34, Q9NVX4, Q9P0R0 | Gene names | ZC3HC1, NIPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (hNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.029209 (rank : 43) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.028581 (rank : 44) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
DOK2_MOUSE
|
||||||
| NC score | 0.025948 (rank : 45) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
CU124_HUMAN
|
||||||
| NC score | 0.024123 (rank : 46) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HW9 | Gene names | C21orf124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf124. | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.022669 (rank : 47) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
YPEL4_HUMAN
|
||||||
| NC score | 0.020131 (rank : 48) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NS1, Q65Z98 | Gene names | YPEL4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein yippee-like 4. | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.019651 (rank : 49) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
FOXL1_MOUSE
|
||||||
| NC score | 0.017825 (rank : 50) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.017699 (rank : 51) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
MADCA_MOUSE
|
||||||
| NC score | 0.015323 (rank : 52) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61826, O35530, Q61278, Q64275, Q8R1M6 | Gene names | Madcam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (mMAdCAM-1). | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.014403 (rank : 53) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.014102 (rank : 54) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.013721 (rank : 55) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.012235 (rank : 56) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.012076 (rank : 57) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.010489 (rank : 58) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.010274 (rank : 59) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
EPHA1_HUMAN
|
||||||
| NC score | 0.009527 (rank : 60) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.009124 (rank : 61) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BEGIN_MOUSE
|
||||||
| NC score | 0.009043 (rank : 62) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CNTN1_HUMAN
|
||||||
| NC score | 0.007241 (rank : 63) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.006751 (rank : 64) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.006040 (rank : 65) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ANKK1_HUMAN
|
||||||
| NC score | 0.002414 (rank : 66) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.002392 (rank : 67) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
HXD1_MOUSE
|
||||||
| NC score | 0.000963 (rank : 68) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||