Please be patient as the page loads
|
PUR2_MOUSE
|
||||||
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PUR2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996569 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
PUR2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
FMT_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 3) | NC score | 0.310260 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
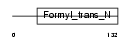
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 4) | NC score | 0.309687 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |
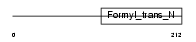
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FTHFD_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 5) | NC score | 0.120609 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
FTHFD_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 6) | NC score | 0.125000 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.119406 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.102666 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.025419 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.006698 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.035356 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.003450 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.010536 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.034090 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.004874 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
DOCK9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.007807 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.043002 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.012812 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
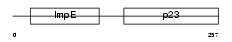
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
PYR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.070447 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
PUR2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64737, Q6NS48 | Gene names | Gart | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
PUR2_HUMAN
|
||||||
| NC score | 0.996569 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
FMT_HUMAN
|
||||||
| NC score | 0.310260 (rank : 3) | θ value | 2.44474e-05 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
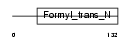
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_MOUSE
|
||||||
| NC score | 0.309687 (rank : 4) | θ value | 2.44474e-05 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FTHFD_MOUSE
|
||||||
| NC score | 0.125000 (rank : 5) | θ value | 9.29e-05 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
FTHFD_HUMAN
|
||||||
| NC score | 0.120609 (rank : 6) | θ value | 7.1131e-05 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.119406 (rank : 7) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.102666 (rank : 8) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.070447 (rank : 9) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.043002 (rank : 10) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.035356 (rank : 11) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.034090 (rank : 12) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.025419 (rank : 13) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.012812 (rank : 14) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.010536 (rank : 15) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
DOCK9_HUMAN
|
||||||
| NC score | 0.007807 (rank : 16) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ29, Q5JUD4, Q5JUD6, Q5T2Q1, Q5TAN8, Q9BZ25, Q9BZ26, Q9BZ27, Q9BZ28, Q9UPU4 | Gene names | DOCK9, KIAA1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.006698 (rank : 17) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.004874 (rank : 18) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.003450 (rank : 19) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||