Please be patient as the page loads
|
NRX1B_HUMAN
|
||||||
| SwissProt Accessions | P58400 | Gene names | NRXN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-beta precursor (Neurexin I-beta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NRX1B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58400 | Gene names | NRXN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-beta precursor (Neurexin I-beta). | |||||
|
NRX1A_HUMAN
|
||||||
| θ value | 8.59026e-160 (rank : 2) | NC score | 0.840307 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_MOUSE
|
||||||
| θ value | 1.41924e-154 (rank : 3) | NC score | 0.844006 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 3.1125e-93 (rank : 4) | NC score | 0.800857 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
NRX3B_HUMAN
|
||||||
| θ value | 1.30768e-91 (rank : 5) | NC score | 0.966111 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HDB5 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-3-beta precursor (Neurexin III-beta). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.16048e-86 (rank : 6) | NC score | 0.790230 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| θ value | 2.47191e-82 (rank : 7) | NC score | 0.839546 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
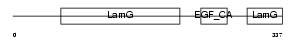
|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 8) | NC score | 0.429288 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.435813 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 10) | NC score | 0.394174 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 11) | NC score | 0.400079 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 12) | NC score | 0.412240 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 13) | NC score | 0.391570 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
FATH_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.109358 (rank : 41) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.213333 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
SLIT1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.122704 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
CSPG4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 17) | NC score | 0.265478 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
SLIT1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.123198 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.204370 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 20) | NC score | 0.147960 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.210816 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.209419 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.154446 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SLIT2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.119252 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1B9, Q9Z166 | Gene names | Slit2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
CDON_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.049993 (rank : 113) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.174514 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
SLIT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.119498 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94813, O95710, Q9Y5Q7 | Gene names | SLIT2, SLIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.409241 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.166355 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.017384 (rank : 117) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.015623 (rank : 118) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
AMOL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.018610 (rank : 116) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.147447 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
ATL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.012407 (rank : 119) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P82987, Q9ULI7 | Gene names | ADAMTSL3, KIAA1233 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 3 precursor (ADAMTSL-3) (Punctin-2). | |||||
|
PRP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.029408 (rank : 114) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.029367 (rank : 115) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
SLIT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.105818 (rank : 44) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.254313 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.051152 (rank : 111) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.057188 (rank : 97) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.084952 (rank : 57) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.088434 (rank : 54) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.093586 (rank : 51) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.086340 (rank : 55) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.091978 (rank : 53) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.095947 (rank : 49) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.134845 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.125486 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.099176 (rank : 47) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.064362 (rank : 83) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.064909 (rank : 82) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052359 (rank : 108) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.058560 (rank : 95) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.215452 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.052894 (rank : 106) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.056526 (rank : 99) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.068891 (rank : 75) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.072193 (rank : 64) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.075025 (rank : 62) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.072068 (rank : 65) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.070666 (rank : 70) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.070675 (rank : 69) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.063623 (rank : 84) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
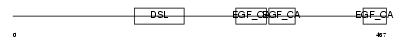
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.071402 (rank : 68) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.071965 (rank : 66) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DNER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.073074 (rank : 63) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.092835 (rank : 52) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
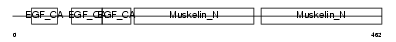
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.093898 (rank : 50) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
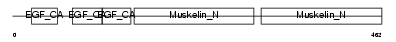
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.070318 (rank : 71) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.068106 (rank : 77) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.075632 (rank : 60) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
GAS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.067075 (rank : 80) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14393, Q7Z7N3 | Gene names | GAS6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
GAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.069361 (rank : 73) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61592, Q99K57 | Gene names | Gas6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
GLPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.085603 (rank : 56) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |
|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.067909 (rank : 78) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.068272 (rank : 76) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061239 (rank : 89) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.061609 (rank : 88) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.149492 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.099080 (rank : 48) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.101625 (rank : 46) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.051622 (rank : 110) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052931 (rank : 105) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050136 (rank : 112) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.063550 (rank : 85) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.061901 (rank : 87) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MFGM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.081175 (rank : 59) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
MFGM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.108804 (rank : 42) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.057801 (rank : 96) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.058601 (rank : 94) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051869 (rank : 109) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.053148 (rank : 104) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.054315 (rank : 101) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.057089 (rank : 98) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.053369 (rank : 102) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053194 (rank : 103) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PGH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.109646 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23219, Q15122 | Gene names | PTGS1, COX1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.106448 (rank : 43) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22437 | Gene names | Ptgs1, Cox-1, Cox1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.125856 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35354, Q16876 | Gene names | PTGS2, COX2 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II). | |||||
|
PGH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.125671 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05769 | Gene names | Ptgs2, Cox-2, Cox2, Pghs-b, Tis10 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II) (Glucocorticoid- regulated inflammatory cyclooxygenase) (Gripghs) (TIS10 protein) (Macrophage activation-associated marker protein P71/73) (PES-2). | |||||
|
PROZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.052809 (rank : 107) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
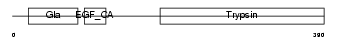
PROZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058983 (rank : 91) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.058944 (rank : 92) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.069849 (rank : 72) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
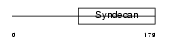
SDC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.066093 (rank : 81) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31431, O00773, Q16833 | Gene names | SDC4 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-4 precursor (SYND4) (Amphiglycan) (Ryudocan core protein). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054985 (rank : 100) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.060315 (rank : 90) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.062159 (rank : 86) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.058645 (rank : 93) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
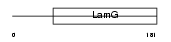
SHBG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.140748 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |
|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
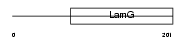
SHBG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.120202 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97497 | Gene names | Shbg | |||
|
Domain Architecture |
|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP). | |||||
|
SLIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.103521 (rank : 45) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.084218 (rank : 58) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.071763 (rank : 67) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
USH2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.140118 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.122417 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.067182 (rank : 79) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.068961 (rank : 74) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.075604 (rank : 61) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
NRX1B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58400 | Gene names | NRXN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-beta precursor (Neurexin I-beta). | |||||
|
NRX3B_HUMAN
|
||||||
| NC score | 0.966111 (rank : 2) | θ value | 1.30768e-91 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HDB5 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-3-beta precursor (Neurexin III-beta). | |||||
|
NRX1A_MOUSE
|
||||||
| NC score | 0.844006 (rank : 3) | θ value | 1.41924e-154 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_HUMAN
|
||||||
| NC score | 0.840307 (rank : 4) | θ value | 8.59026e-160 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| NC score | 0.839546 (rank : 5) | θ value | 2.47191e-82 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.800857 (rank : 6) | θ value | 3.1125e-93 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.790230 (rank : 7) | θ value | 2.16048e-86 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.435813 (rank : 8) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.429288 (rank : 9) | θ value | 1.99992e-07 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.412240 (rank : 10) | θ value | 1.87187e-05 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.409241 (rank : 11) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.400079 (rank : 12) | θ value | 1.09739e-05 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.394174 (rank : 13) | θ value | 4.92598e-06 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.391570 (rank : 14) | θ value | 3.19293e-05 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CSPG4_MOUSE
|
||||||
| NC score | 0.265478 (rank : 15) | θ value | 0.00228821 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHY0, Q5DTG1, Q8BPI8, Q8CE79 | Gene names | Cspg4, An2, Kiaa4232, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (AN2 proteoglycan). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.254313 (rank : 16) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CSPG4_HUMAN
|
||||||
| NC score | 0.215452 (rank : 17) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.213333 (rank : 18) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.210816 (rank : 19) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.209419 (rank : 20) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.204370 (rank : 21) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.174514 (rank : 22) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.166355 (rank : 23) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.154446 (rank : 24) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.149492 (rank : 25) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.147960 (rank : 26) | θ value | 0.00509761 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.147447 (rank : 27) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SHBG_HUMAN
|
||||||
| NC score | 0.140748 (rank : 28) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |

|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.140118 (rank : 29) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.134845 (rank : 30) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
PGH2_HUMAN
|
||||||
| NC score | 0.125856 (rank : 31) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35354, Q16876 | Gene names | PTGS2, COX2 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II). | |||||
|
PGH2_MOUSE
|
||||||
| NC score | 0.125671 (rank : 32) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05769 | Gene names | Ptgs2, Cox-2, Cox2, Pghs-b, Tis10 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II) (Glucocorticoid- regulated inflammatory cyclooxygenase) (Gripghs) (TIS10 protein) (Macrophage activation-associated marker protein P71/73) (PES-2). | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.125486 (rank : 33) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
SLIT1_HUMAN
|
||||||
| NC score | 0.123198 (rank : 34) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
SLIT1_MOUSE
|
||||||
| NC score | 0.122704 (rank : 35) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.122417 (rank : 36) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
SHBG_MOUSE
|
||||||
| NC score | 0.120202 (rank : 37) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97497 | Gene names | Shbg | |||
|
Domain Architecture |

|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP). | |||||
|
SLIT2_HUMAN
|
||||||
| NC score | 0.119498 (rank : 38) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94813, O95710, Q9Y5Q7 | Gene names | SLIT2, SLIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
SLIT2_MOUSE
|
||||||
| NC score | 0.119252 (rank : 39) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1B9, Q9Z166 | Gene names | Slit2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
PGH1_HUMAN
|
||||||
| NC score | 0.109646 (rank : 40) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23219, Q15122 | Gene names | PTGS1, COX1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
FATH_HUMAN
|
||||||
| NC score | 0.109358 (rank : 41) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.108804 (rank : 42) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
PGH1_MOUSE
|
||||||
| NC score | 0.106448 (rank : 43) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22437 | Gene names | Ptgs1, Cox-1, Cox1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
SLIT3_MOUSE
|
||||||
| NC score | 0.105818 (rank : 44) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
SLIT3_HUMAN
|
||||||
| NC score | 0.103521 (rank : 45) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.101625 (rank : 46) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.099176 (rank : 47) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.099080 (rank : 48) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.095947 (rank : 49) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.093898 (rank : 50) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.093586 (rank : 51) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.092835 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.091978 (rank : 53) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.088434 (rank : 54) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.086340 (rank : 55) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
GLPC_HUMAN
|
||||||
| NC score | 0.085603 (rank : 56) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.084952 (rank : 57) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.084218 (rank : 58) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.081175 (rank : 59) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.075632 (rank : 60) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.075604 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.075025 (rank : 62) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DNER_MOUSE
|
||||||
| NC score | 0.073074 (rank : 63) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.072193 (rank : 64) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.072068 (rank : 65) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.071965 (rank : 66) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.071763 (rank : 67) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.071402 (rank : 68) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.070675 (rank : 69) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.070666 (rank : 70) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.070318 (rank : 71) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.069849 (rank : 72) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
GAS6_MOUSE
|
||||||
| NC score | 0.069361 (rank : 73) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61592, Q99K57 | Gene names | Gas6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.068961 (rank : 74) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.068891 (rank : 75) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.068272 (rank : 76) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.068106 (rank : 77) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.067909 (rank : 78) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.067182 (rank : 79) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
GAS6_HUMAN
|
||||||
| NC score | 0.067075 (rank : 80) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14393, Q7Z7N3 | Gene names | GAS6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
SDC4_HUMAN
|
||||||
| NC score | 0.066093 (rank : 81) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31431, O00773, Q16833 | Gene names | SDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-4 precursor (SYND4) (Amphiglycan) (Ryudocan core protein). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.064909 (rank : 82) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.064362 (rank : 83) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.063623 (rank : 84) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.063550 (rank : 85) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.062159 (rank : 86) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.061901 (rank : 87) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.061609 (rank : 88) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.061239 (rank : 89) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.060315 (rank : 90) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
PROZ_MOUSE
|
||||||
| NC score | 0.058983 (rank : 91) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.058944 (rank : 92) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.058645 (rank : 93) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.058601 (rank : 94) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.058560 (rank : 95) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.057801 (rank : 96) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.057188 (rank : 97) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.057089 (rank : 98) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.056526 (rank : 99) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.054985 (rank : 100) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.054315 (rank : 101) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.053369 (rank : 102) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.053194 (rank : 103) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.053148 (rank : 104) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.052931 (rank : 105) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.052894 (rank : 106) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
PROZ_HUMAN
|
||||||
| NC score | 0.052809 (rank : 107) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.052359 (rank : 108) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.051869 (rank : 109) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.051622 (rank : 110) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.051152 (rank : 111) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.050136 (rank : 112) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
CDON_HUMAN
|
||||||
| NC score | 0.049993 (rank : 113) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
PRP6_HUMAN
|
||||||
| NC score | 0.029408 (rank : 114) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| NC score | 0.029367 (rank : 115) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
AMOL2_HUMAN
|
||||||
| NC score | 0.018610 (rank : 116) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.017384 (rank : 117) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.015623 (rank : 118) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
ATL3_HUMAN
|
||||||
| NC score | 0.012407 (rank : 119) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P82987, Q9ULI7 | Gene names | ADAMTSL3, KIAA1233 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 3 precursor (ADAMTSL-3) (Punctin-2). | |||||