Please be patient as the page loads
|
MO4L2_HUMAN
|
||||||
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MO4L2_HUMAN
|
||||||
| θ value | 3.04821e-165 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 6.1562e-158 (rank : 2) | NC score | 0.985386 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MO4L1_HUMAN
|
||||||
| θ value | 4.93473e-115 (rank : 3) | NC score | 0.961752 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |
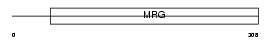
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MO4L1_MOUSE
|
||||||
| θ value | 4.93473e-115 (rank : 4) | NC score | 0.961466 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |
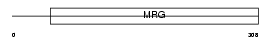
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MORF4_HUMAN
|
||||||
| θ value | 1.43912e-106 (rank : 5) | NC score | 0.976190 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |
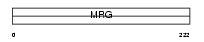
|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 6) | NC score | 0.445337 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 7) | NC score | 0.427040 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.093920 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.064519 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.091941 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MRP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.101566 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.088247 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.025799 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.080554 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.024706 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.013454 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
LMX1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.024251 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |
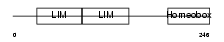
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
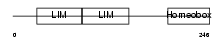
LMX1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.024184 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.012485 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.050793 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.015616 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.031912 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
ING5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.030030 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.013594 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.030024 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.025839 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.035760 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
WFS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.056089 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
SORC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.021337 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.044731 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.024597 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
SYTC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.037571 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.013288 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.028600 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.036281 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.020092 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.025089 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.071960 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.032980 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.024685 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025948 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
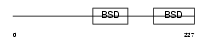
TF2H1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.023750 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32780, Q9H5K5, Q9NQD9 | Gene names | GTF2H1, BTF2 | |||
|
Domain Architecture |
|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.000736 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.017695 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.020389 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.027710 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
FUS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.026802 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030118 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.044726 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.016934 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.021866 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.010403 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
IF5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.080462 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.033646 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.038333 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.018093 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
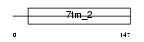
GP123_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.013990 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
H14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.027232 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
IPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.029171 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.023627 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
PRR6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.019698 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXS4, Q5SX21, Q6PIA5 | Gene names | Prr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6. | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.031968 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SYEP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.028307 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.011612 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.033897 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.018582 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.023259 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012763 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.015643 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.010943 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.014714 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
IF2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.041955 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
IF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.060129 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
MARK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | -0.002109 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.044190 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.023572 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.010289 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.032006 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.015365 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.012830 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.015001 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.003799 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.020090 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.013745 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SPC25_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.009774 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.023509 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.017379 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003788 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.015335 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
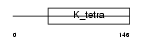
ZN297_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | -0.000998 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15209, Q5STL0, Q5STR7, Q8WV82 | Gene names | ZNF297, BING1, ZBTB22A | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 297 (BING1 protein) (Zinc finger and BTB domain- containing protein 22A). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.067267 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SEC62_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050593 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
MO4L2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.04821e-165 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.985386 (rank : 2) | θ value | 6.1562e-158 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MORF4_HUMAN
|
||||||
| NC score | 0.976190 (rank : 3) | θ value | 1.43912e-106 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MO4L1_HUMAN
|
||||||
| NC score | 0.961752 (rank : 4) | θ value | 4.93473e-115 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MO4L1_MOUSE
|
||||||
| NC score | 0.961466 (rank : 5) | θ value | 4.93473e-115 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.445337 (rank : 6) | θ value | 3.77169e-06 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.427040 (rank : 7) | θ value | 3.77169e-06 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.101566 (rank : 8) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.093920 (rank : 9) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.091941 (rank : 10) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.088247 (rank : 11) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.080554 (rank : 12) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
IF5_MOUSE
|
||||||
| NC score | 0.080462 (rank : 13) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.071960 (rank : 14) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.067267 (rank : 15) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.064519 (rank : 16) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
IF5_HUMAN
|
||||||
| NC score | 0.060129 (rank : 17) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
WFS1_HUMAN
|
||||||
| NC score | 0.056089 (rank : 18) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.050793 (rank : 19) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.050593 (rank : 20) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.044731 (rank : 21) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.044726 (rank : 22) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.044190 (rank : 23) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
IF2B_HUMAN
|
||||||
| NC score | 0.041955 (rank : 24) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.038333 (rank : 25) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SYTC_MOUSE
|
||||||
| NC score | 0.037571 (rank : 26) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.036281 (rank : 27) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.035760 (rank : 28) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.033897 (rank : 29) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.033646 (rank : 30) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.032980 (rank : 31) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.032006 (rank : 32) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.031968 (rank : 33) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.031912 (rank : 34) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.030118 (rank : 35) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ING5_MOUSE
|
||||||
| NC score | 0.030030 (rank : 36) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.030024 (rank : 37) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
IPP1_MOUSE
|
||||||
| NC score | 0.029171 (rank : 38) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.028600 (rank : 39) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.028307 (rank : 40) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.027710 (rank : 41) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.027232 (rank : 42) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.026802 (rank : 43) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.025948 (rank : 44) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.025839 (rank : 45) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.025799 (rank : 46) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.025089 (rank : 47) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.024706 (rank : 48) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.024685 (rank : 49) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SYN3_MOUSE
|
||||||
| NC score | 0.024597 (rank : 50) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
LMX1B_HUMAN
|
||||||
| NC score | 0.024251 (rank : 51) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LMX1B_MOUSE
|
||||||
| NC score | 0.024184 (rank : 52) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
TF2H1_HUMAN
|
||||||
| NC score | 0.023750 (rank : 53) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32780, Q9H5K5, Q9NQD9 | Gene names | GTF2H1, BTF2 | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.023627 (rank : 54) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.023572 (rank : 55) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.023509 (rank : 56) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.023259 (rank : 57) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.021866 (rank : 58) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
SORC2_MOUSE
|
||||||
| NC score | 0.021337 (rank : 59) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.020389 (rank : 60) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.020092 (rank : 61) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.020090 (rank : 62) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PRR6_MOUSE
|
||||||
| NC score | 0.019698 (rank : 63) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXS4, Q5SX21, Q6PIA5 | Gene names | Prr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6. | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.018582 (rank : 64) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.018093 (rank : 65) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.017695 (rank : 66) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.017379 (rank : 67) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.016934 (rank : 68) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.015643 (rank : 69) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.015616 (rank : 70) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.015365 (rank : 71) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.015335 (rank : 72) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.015001 (rank : 73) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.014714 (rank : 74) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
GP123_HUMAN
|
||||||
| NC score | 0.013990 (rank : 75) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.013745 (rank : 76) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.013594 (rank : 77) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.013454 (rank : 78) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.013288 (rank : 79) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.012830 (rank : 80) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.012763 (rank : 81) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.012485 (rank : 82) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.011612 (rank : 83) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.010943 (rank : 84) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.010403 (rank : 85) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.010289 (rank : 86) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SPC25_MOUSE
|
||||||
| NC score | 0.009774 (rank : 87) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.003799 (rank : 88) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.003788 (rank : 89) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.000736 (rank : 90) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN297_HUMAN
|
||||||
| NC score | -0.000998 (rank : 91) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15209, Q5STL0, Q5STR7, Q8WV82 | Gene names | ZNF297, BING1, ZBTB22A | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297 (BING1 protein) (Zinc finger and BTB domain- containing protein 22A). | |||||
|
MARK4_HUMAN
|
||||||
| NC score | -0.002109 (rank : 92) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||