Please be patient as the page loads
|
MO4L1_MOUSE
|
||||||
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |
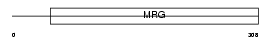
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MO4L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999728 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |
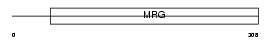
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MO4L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MORF4_HUMAN
|
||||||
| θ value | 1.38424e-125 (rank : 3) | NC score | 0.982180 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |
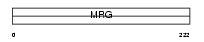
|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MO4L2_HUMAN
|
||||||
| θ value | 4.93473e-115 (rank : 4) | NC score | 0.961466 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 1.13764e-111 (rank : 5) | NC score | 0.958993 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 6) | NC score | 0.539503 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 7) | NC score | 0.521892 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.084029 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.090954 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.068241 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.028857 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
SEC62_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.091450 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.032360 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SEC62_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.092793 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.041717 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
IF5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.094134 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.048818 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.032758 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.031480 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.021338 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.012474 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HDAC9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.024507 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.003370 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.031709 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
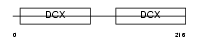
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.060598 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.021288 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.027825 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.020197 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.023887 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
HDAC9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.032544 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
IF5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.073522 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.021896 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
IF2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.047338 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.024672 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.025833 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.017563 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DLGP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.014815 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
K1199_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.018906 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUJ3, Q9H1K5, Q9NPN9, Q9ULM1 | Gene names | KIAA1199 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1199 precursor. | |||||
|
MMTA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.031598 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.033753 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.009563 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.008642 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
FGF18_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.012109 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76093, Q6UWF1 | Gene names | FGF18 | |||
|
Domain Architecture |
|
|||||
| Description | Fibroblast growth factor 18 precursor (FGF-18) (zFGF5). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.015007 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.038810 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.021273 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.021243 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.009024 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
LYAR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.017483 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.010146 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.000924 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.055240 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
MO4L1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MO4L1_HUMAN
|
||||||
| NC score | 0.999728 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MORF4_HUMAN
|
||||||
| NC score | 0.982180 (rank : 3) | θ value | 1.38424e-125 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MO4L2_HUMAN
|
||||||
| NC score | 0.961466 (rank : 4) | θ value | 4.93473e-115 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.958993 (rank : 5) | θ value | 1.13764e-111 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.539503 (rank : 6) | θ value | 2.35696e-16 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.521892 (rank : 7) | θ value | 5.8054e-15 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
IF5_MOUSE
|
||||||
| NC score | 0.094134 (rank : 8) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.092793 (rank : 9) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
SEC62_HUMAN
|
||||||
| NC score | 0.091450 (rank : 10) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.090954 (rank : 11) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.084029 (rank : 12) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
IF5_HUMAN
|
||||||
| NC score | 0.073522 (rank : 13) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.068241 (rank : 14) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.060598 (rank : 15) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.055240 (rank : 16) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.048818 (rank : 17) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
IF2B_HUMAN
|
||||||
| NC score | 0.047338 (rank : 18) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.041717 (rank : 19) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.038810 (rank : 20) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.033753 (rank : 21) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.032758 (rank : 22) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
HDAC9_MOUSE
|
||||||
| NC score | 0.032544 (rank : 23) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.032360 (rank : 24) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.031709 (rank : 25) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
MMTA2_MOUSE
|
||||||
| NC score | 0.031598 (rank : 26) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LX5, Q9CSK6 | Gene names | Mmtag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 homolog. | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.031480 (rank : 27) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.028857 (rank : 28) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.027825 (rank : 29) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.025833 (rank : 30) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.024672 (rank : 31) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
HDAC9_HUMAN
|
||||||
| NC score | 0.024507 (rank : 32) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.023887 (rank : 33) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.021896 (rank : 34) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.021338 (rank : 35) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.021288 (rank : 36) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.021273 (rank : 37) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.021243 (rank : 38) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.020197 (rank : 39) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
K1199_HUMAN
|
||||||
| NC score | 0.018906 (rank : 40) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUJ3, Q9H1K5, Q9NPN9, Q9ULM1 | Gene names | KIAA1199 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1199 precursor. | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.017563 (rank : 41) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
LYAR_HUMAN
|
||||||
| NC score | 0.017483 (rank : 42) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.015007 (rank : 43) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DLGP1_MOUSE
|
||||||
| NC score | 0.014815 (rank : 44) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.012474 (rank : 45) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FGF18_HUMAN
|
||||||
| NC score | 0.012109 (rank : 46) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76093, Q6UWF1 | Gene names | FGF18 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor 18 precursor (FGF-18) (zFGF5). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.010146 (rank : 47) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.009563 (rank : 48) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.009024 (rank : 49) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.008642 (rank : 50) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.003370 (rank : 51) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.000924 (rank : 52) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||