Please be patient as the page loads
|
MBD2_HUMAN
|
||||||
| SwissProt Accessions | Q9UBB5, O95242, Q9UIS8 | Gene names | MBD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MBD2_HUMAN
|
||||||
| θ value | 2.67656e-153 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UBB5, O95242, Q9UIS8 | Gene names | MBD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase). | |||||
|
MBD2_MOUSE
|
||||||
| θ value | 1.46868e-151 (rank : 2) | NC score | 0.995763 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2E1, Q811D9, Q9Z2D9 | Gene names | Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2). | |||||
|
MBD3_HUMAN
|
||||||
| θ value | 3.31004e-111 (rank : 3) | NC score | 0.986145 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95983, Q6PIL9, Q6PJZ9, Q86XF4 | Gene names | MBD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3 (Methyl-CpG-binding protein MBD3). | |||||
|
MBD3_MOUSE
|
||||||
| θ value | 1.64275e-110 (rank : 4) | NC score | 0.985775 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2D8, Q792D3, Q8CFJ1 | Gene names | Mbd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3 (Methyl-CpG-binding protein MBD3). | |||||
|
MB3L1_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 5) | NC score | 0.832795 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWY6 | Gene names | MBD3L1, MBD3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 1 (MBD3-like 1). | |||||
|
MB3L1_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 6) | NC score | 0.784013 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D9H3, Q6P8W5, Q9D2S3 | Gene names | Mbd3l1, Mbd3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 1 (MBD3-like 1). | |||||
|
MBD1_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 7) | NC score | 0.539285 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
MBD1_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 8) | NC score | 0.532367 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIS9, O15248, O95241, Q7Z7B5, Q8N4W4, Q9UNZ6, Q9UNZ7, Q9UNZ8, Q9UNZ9 | Gene names | MBD1, PCM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1) (Protein containing methyl-CpG-binding domain 1). | |||||
|
MB3L2_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 9) | NC score | 0.720573 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 10) | NC score | 0.508760 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MECP2_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 11) | NC score | 0.502581 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MBD4_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 12) | NC score | 0.376482 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2D7, Q792D2, Q8R3R3 | Gene names | Mbd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Mismatch-specific DNA N-glycosylase). | |||||
|
MBD4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 13) | NC score | 0.368141 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95243, Q7Z4T3, Q96F09 | Gene names | MBD4, MED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Methyl-CpG-binding endonuclease 1) (Mismatch-specific DNA N-glycosylase). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 14) | NC score | 0.042308 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.018894 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.086071 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.074969 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.074892 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.024420 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.021412 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.023443 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
ZN148_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.005945 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.012054 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NR2E3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.005913 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |
|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
THIKA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.018272 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921H8 | Gene names | Acaa1a, Acaa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketoacyl-CoA thiolase A, peroxisomal precursor (EC 2.3.1.16) (Beta- ketothiolase A) (Acetyl-CoA acyltransferase A) (Peroxisomal 3-oxoacyl- CoA thiolase A). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.012736 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.023255 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.028657 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.019114 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN148_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.004583 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
A20A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.015124 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 698 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TYW2, Q9H0H6 | Gene names | ANKRD20A1, ANKRD20A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A1. | |||||
|
A20A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.015470 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q4UJ75 | Gene names | ANKRD20A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A2. | |||||
|
A20A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.014930 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VUR7 | Gene names | ANKRD20A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A3. | |||||
|
A20A4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.015059 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SQ80 | Gene names | ANKRD20A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A4. | |||||
|
CN104_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.033938 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BPI1, Q9CZC7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf104 homolog. | |||||
|
HEM6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.016984 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36551, Q14060, Q8IZ45, Q96AF3 | Gene names | CPOX, CPO, CPX | |||
|
Domain Architecture |

|
|||||
| Description | Coproporphyrinogen III oxidase, mitochondrial precursor (EC 1.3.3.3) (Coproporphyrinogenase) (Coprogen oxidase) (COX). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.015721 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.016596 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.011999 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.013976 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
PODO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.009619 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP85, Q8N6Q5 | Gene names | NPHS2 | |||
|
Domain Architecture |
|
|||||
| Description | Podocin. | |||||
|
P80C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.023327 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | -0.001371 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SURF6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.015236 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |
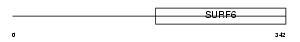
|
|||||
| Description | Surfeit locus protein 6. | |||||
|
TNK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | -0.001207 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
TOP2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.007868 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
CXCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.051168 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
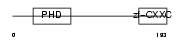
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.051343 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
MBD2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.67656e-153 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UBB5, O95242, Q9UIS8 | Gene names | MBD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2) (Demethylase) (DMTase). | |||||
|
MBD2_MOUSE
|
||||||
| NC score | 0.995763 (rank : 2) | θ value | 1.46868e-151 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2E1, Q811D9, Q9Z2D9 | Gene names | Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2). | |||||
|
MBD3_HUMAN
|
||||||
| NC score | 0.986145 (rank : 3) | θ value | 3.31004e-111 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95983, Q6PIL9, Q6PJZ9, Q86XF4 | Gene names | MBD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3 (Methyl-CpG-binding protein MBD3). | |||||
|
MBD3_MOUSE
|
||||||
| NC score | 0.985775 (rank : 4) | θ value | 1.64275e-110 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2D8, Q792D3, Q8CFJ1 | Gene names | Mbd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3 (Methyl-CpG-binding protein MBD3). | |||||
|
MB3L1_HUMAN
|
||||||
| NC score | 0.832795 (rank : 5) | θ value | 3.3877e-31 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWY6 | Gene names | MBD3L1, MBD3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 1 (MBD3-like 1). | |||||
|
MB3L1_MOUSE
|
||||||
| NC score | 0.784013 (rank : 6) | θ value | 2.27762e-19 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D9H3, Q6P8W5, Q9D2S3 | Gene names | Mbd3l1, Mbd3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 1 (MBD3-like 1). | |||||
|
MB3L2_HUMAN
|
||||||
| NC score | 0.720573 (rank : 7) | θ value | 7.58209e-15 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
MBD1_MOUSE
|
||||||
| NC score | 0.539285 (rank : 8) | θ value | 3.63628e-17 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
MBD1_HUMAN
|
||||||
| NC score | 0.532367 (rank : 9) | θ value | 1.38178e-16 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIS9, O15248, O95241, Q7Z7B5, Q8N4W4, Q9UNZ6, Q9UNZ7, Q9UNZ8, Q9UNZ9 | Gene names | MBD1, PCM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1) (Protein containing methyl-CpG-binding domain 1). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.508760 (rank : 10) | θ value | 1.133e-10 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MECP2_MOUSE
|
||||||
| NC score | 0.502581 (rank : 11) | θ value | 1.47974e-10 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MBD4_MOUSE
|
||||||
| NC score | 0.376482 (rank : 12) | θ value | 6.43352e-06 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2D7, Q792D2, Q8R3R3 | Gene names | Mbd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Mismatch-specific DNA N-glycosylase). | |||||
|
MBD4_HUMAN
|
||||||
| NC score | 0.368141 (rank : 13) | θ value | 1.87187e-05 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95243, Q7Z4T3, Q96F09 | Gene names | MBD4, MED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Methyl-CpG-binding endonuclease 1) (Mismatch-specific DNA N-glycosylase). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.086071 (rank : 14) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.074969 (rank : 15) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.074892 (rank : 16) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
CXCC1_MOUSE
|
||||||
| NC score | 0.051343 (rank : 17) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.051168 (rank : 18) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.042308 (rank : 19) | θ value | 3.19293e-05 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
CN104_MOUSE
|
||||||
| NC score | 0.033938 (rank : 20) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BPI1, Q9CZC7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf104 homolog. | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.028657 (rank : 21) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.024420 (rank : 22) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.023443 (rank : 23) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.023327 (rank : 24) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.023255 (rank : 25) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.021412 (rank : 26) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.019114 (rank : 27) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.018894 (rank : 28) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
THIKA_MOUSE
|
||||||
| NC score | 0.018272 (rank : 29) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921H8 | Gene names | Acaa1a, Acaa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketoacyl-CoA thiolase A, peroxisomal precursor (EC 2.3.1.16) (Beta- ketothiolase A) (Acetyl-CoA acyltransferase A) (Peroxisomal 3-oxoacyl- CoA thiolase A). | |||||
|
HEM6_HUMAN
|
||||||
| NC score | 0.016984 (rank : 30) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36551, Q14060, Q8IZ45, Q96AF3 | Gene names | CPOX, CPO, CPX | |||
|
Domain Architecture |

|
|||||
| Description | Coproporphyrinogen III oxidase, mitochondrial precursor (EC 1.3.3.3) (Coproporphyrinogenase) (Coprogen oxidase) (COX). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.016596 (rank : 31) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.015721 (rank : 32) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
A20A2_HUMAN
|
||||||
| NC score | 0.015470 (rank : 33) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q4UJ75 | Gene names | ANKRD20A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A2. | |||||
|
SURF6_MOUSE
|
||||||
| NC score | 0.015236 (rank : 34) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
A20A1_HUMAN
|
||||||
| NC score | 0.015124 (rank : 35) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 698 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TYW2, Q9H0H6 | Gene names | ANKRD20A1, ANKRD20A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A1. | |||||
|
A20A4_HUMAN
|
||||||
| NC score | 0.015059 (rank : 36) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SQ80 | Gene names | ANKRD20A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A4. | |||||
|
A20A3_HUMAN
|
||||||
| NC score | 0.014930 (rank : 37) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VUR7 | Gene names | ANKRD20A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A3. | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.013976 (rank : 38) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.012736 (rank : 39) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.012054 (rank : 40) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.011999 (rank : 41) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
PODO_HUMAN
|
||||||
| NC score | 0.009619 (rank : 42) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP85, Q8N6Q5 | Gene names | NPHS2 | |||
|
Domain Architecture |

|
|||||
| Description | Podocin. | |||||
|
TOP2B_HUMAN
|
||||||
| NC score | 0.007868 (rank : 43) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
ZN148_HUMAN
|
||||||
| NC score | 0.005945 (rank : 44) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
NR2E3_MOUSE
|
||||||
| NC score | 0.005913 (rank : 45) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |

|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
ZN148_MOUSE
|
||||||
| NC score | 0.004583 (rank : 46) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
TNK1_HUMAN
|
||||||
| NC score | -0.001207 (rank : 47) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | -0.001371 (rank : 48) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||