Please be patient as the page loads
|
IF3X_HUMAN
|
||||||
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IF3X_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 2) | NC score | 0.490444 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 3) | NC score | 0.484287 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 4) | NC score | 0.478159 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
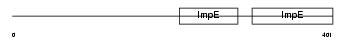
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 5) | NC score | 0.477630 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.458000 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.499901 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 8) | NC score | 0.502521 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 9) | NC score | 0.481883 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
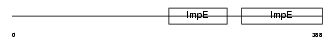
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 10) | NC score | 0.478339 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.456289 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
APBP2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.365787 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
APBP2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 13) | NC score | 0.365795 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
TTC19_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.340349 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC19_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.317232 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
K0423_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.058789 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
ANGP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.030117 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.044585 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.044032 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.070013 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
U383_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.058005 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYL2 | Gene names | C4orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
ANGP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.027203 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.022061 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
M4K1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.005588 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92918 | Gene names | MAP4K1, HPK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 1 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 1) (MEK kinase kinase 1) (MEKKK 1) (Hematopoietic progenitor kinase). | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.040840 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
EXOC5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.047561 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TPX4 | Gene names | Exoc5, Sec10l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.044040 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
M3K5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.004374 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
PCD20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.011486 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
PCD20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.011546 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.031262 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
ECT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.017022 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.032667 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.033515 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.012404 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
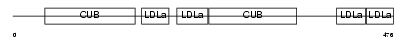
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
PHF22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.051810 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
XRCC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.026360 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |
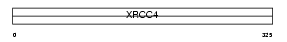
|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
EXOC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.038521 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00471 | Gene names | EXOC5, SEC10, SEC10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10) (hSec10). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.015673 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
AQP12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.018192 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHJ2 | Gene names | Aqp12a, Aqp12 | |||
|
Domain Architecture |

|
|||||
| Description | Aquaporin-12A (AQP-12). | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.009457 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.009306 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.009349 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
TTC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.026591 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.062117 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.060678 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.125641 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.073722 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.068490 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.070424 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.069370 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
IF3X_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.502521 (rank : 2) | θ value | 1.17247e-07 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.499901 (rank : 3) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.490444 (rank : 4) | θ value | 5.08577e-11 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.484287 (rank : 5) | θ value | 1.47974e-10 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.481883 (rank : 6) | θ value | 5.81887e-07 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.478339 (rank : 7) | θ value | 1.29631e-06 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.478159 (rank : 8) | θ value | 3.64472e-09 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.477630 (rank : 9) | θ value | 4.76016e-09 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.458000 (rank : 10) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC1_MOUSE
|
||||||
| NC score | 0.456289 (rank : 11) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
APBP2_MOUSE
|
||||||
| NC score | 0.365795 (rank : 12) | θ value | 0.00035302 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
APBP2_HUMAN
|
||||||
| NC score | 0.365787 (rank : 13) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
TTC19_HUMAN
|
||||||
| NC score | 0.340349 (rank : 14) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC19_MOUSE
|
||||||
| NC score | 0.317232 (rank : 15) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.125641 (rank : 16) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.073722 (rank : 17) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.070424 (rank : 18) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.070013 (rank : 19) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.069370 (rank : 20) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.068490 (rank : 21) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.062117 (rank : 22) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.060678 (rank : 23) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.058789 (rank : 24) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
U383_HUMAN
|
||||||
| NC score | 0.058005 (rank : 25) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYL2 | Gene names | C4orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
PHF22_HUMAN
|
||||||
| NC score | 0.051810 (rank : 26) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
EXOC5_MOUSE
|
||||||
| NC score | 0.047561 (rank : 27) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TPX4 | Gene names | Exoc5, Sec10l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.044585 (rank : 28) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.044040 (rank : 29) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.044032 (rank : 30) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.040840 (rank : 31) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
EXOC5_HUMAN
|
||||||
| NC score | 0.038521 (rank : 32) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00471 | Gene names | EXOC5, SEC10, SEC10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10) (hSec10). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.033515 (rank : 33) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.032667 (rank : 34) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.031262 (rank : 35) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
ANGP1_MOUSE
|
||||||
| NC score | 0.030117 (rank : 36) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP1_HUMAN
|
||||||
| NC score | 0.027203 (rank : 37) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.026591 (rank : 38) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
XRCC4_MOUSE
|
||||||
| NC score | 0.026360 (rank : 39) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924T3, Q8BKC9, Q8BU02, Q9D6U6 | Gene names | Xrcc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.022061 (rank : 40) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
AQP12_MOUSE
|
||||||
| NC score | 0.018192 (rank : 41) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHJ2 | Gene names | Aqp12a, Aqp12 | |||
|
Domain Architecture |

|
|||||
| Description | Aquaporin-12A (AQP-12). | |||||
|
ECT2_MOUSE
|
||||||
| NC score | 0.017022 (rank : 42) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07139 | Gene names | Ect2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.015673 (rank : 43) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.012404 (rank : 44) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
PCD20_MOUSE
|
||||||
| NC score | 0.011546 (rank : 45) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
PCD20_HUMAN
|
||||||
| NC score | 0.011486 (rank : 46) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.009457 (rank : 47) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.009349 (rank : 48) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.009306 (rank : 49) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
M4K1_HUMAN
|
||||||
| NC score | 0.005588 (rank : 50) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92918 | Gene names | MAP4K1, HPK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 1 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 1) (MEK kinase kinase 1) (MEKKK 1) (Hematopoietic progenitor kinase). | |||||
|
M3K5_MOUSE
|
||||||
| NC score | 0.004374 (rank : 51) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||