Please be patient as the page loads
|
APBP2_MOUSE
|
||||||
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APBP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999892 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
APBP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 3) | NC score | 0.490368 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 4) | NC score | 0.481994 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 5) | NC score | 0.445669 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC1_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 6) | NC score | 0.455041 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.457221 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
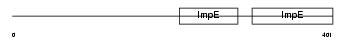
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 8) | NC score | 0.483755 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 9) | NC score | 0.484749 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 10) | NC score | 0.452175 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 11) | NC score | 0.463733 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
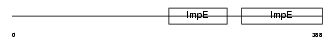
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 12) | NC score | 0.459446 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
IF3X_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 13) | NC score | 0.365795 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.156473 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC19_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.239827 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.141460 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
GPSM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.077595 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.078850 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.028996 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TTC19_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.251298 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
GLTL5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.014305 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.042782 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
S3TC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.038808 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VA5, Q8BWH2, Q8BXB8, Q8C0G6 | Gene names | Sh3tc2, Kiaa1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
MORC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.018798 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VD1, Q7L8E2, Q9NSG7, Q9Y6D4 | Gene names | MORC1, MORC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1. | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.028399 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
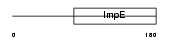
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.034452 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.043885 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
DPEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.009915 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31428 | Gene names | Dpep1, Mbd1, Rdp | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidase 1 precursor (EC 3.4.13.19) (Microsomal dipeptidase) (Renal dipeptidase) (Membrane-bound dipeptidase 1) (MBD-1). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.006381 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.055492 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.054811 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.124777 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.070712 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.065891 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
APBP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
APBP2_HUMAN
|
||||||
| NC score | 0.999892 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.490368 (rank : 3) | θ value | 1.02475e-11 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.484749 (rank : 4) | θ value | 1.17247e-07 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.483755 (rank : 5) | θ value | 5.26297e-08 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.481994 (rank : 6) | θ value | 3.89403e-11 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.463733 (rank : 7) | θ value | 2.61198e-07 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.459446 (rank : 8) | θ value | 5.81887e-07 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.457221 (rank : 9) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC1_MOUSE
|
||||||
| NC score | 0.455041 (rank : 10) | θ value | 1.80886e-08 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.452175 (rank : 11) | θ value | 1.53129e-07 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.445669 (rank : 12) | θ value | 3.64472e-09 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
IF3X_HUMAN
|
||||||
| NC score | 0.365795 (rank : 13) | θ value | 0.00035302 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
TTC19_HUMAN
|
||||||
| NC score | 0.251298 (rank : 14) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC19_MOUSE
|
||||||
| NC score | 0.239827 (rank : 15) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.156473 (rank : 16) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.141460 (rank : 17) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.124777 (rank : 18) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
GPSM1_MOUSE
|
||||||
| NC score | 0.078850 (rank : 19) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_HUMAN
|
||||||
| NC score | 0.077595 (rank : 20) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.070712 (rank : 21) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.065891 (rank : 22) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.055492 (rank : 23) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.054811 (rank : 24) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.043885 (rank : 25) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.042782 (rank : 26) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
S3TC2_MOUSE
|
||||||
| NC score | 0.038808 (rank : 27) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VA5, Q8BWH2, Q8BXB8, Q8C0G6 | Gene names | Sh3tc2, Kiaa1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.034452 (rank : 28) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.028996 (rank : 29) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.028399 (rank : 30) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
MORC1_HUMAN
|
||||||
| NC score | 0.018798 (rank : 31) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86VD1, Q7L8E2, Q9NSG7, Q9Y6D4 | Gene names | MORC1, MORC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 1. | |||||
|
GLTL5_HUMAN
|
||||||
| NC score | 0.014305 (rank : 32) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
DPEP1_MOUSE
|
||||||
| NC score | 0.009915 (rank : 33) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31428 | Gene names | Dpep1, Mbd1, Rdp | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidase 1 precursor (EC 3.4.13.19) (Microsomal dipeptidase) (Renal dipeptidase) (Membrane-bound dipeptidase 1) (MBD-1). | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.006381 (rank : 34) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||