Please be patient as the page loads
|
HEM1_MOUSE
|
||||||
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HEM0_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984095 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
HEM0_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984305 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08680, Q64452, Q9DCN0 | Gene names | Alas2 | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
HEM1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990798 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13196 | Gene names | ALAS1, ALASH | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
HEM1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
KBL_MOUSE
|
||||||
| θ value | 8.28633e-54 (rank : 5) | NC score | 0.893235 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88986 | Gene names | Gcat, Kbl | |||
|
Domain Architecture |
|
|||||
| Description | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial precursor (EC 2.3.1.29) (AKB ligase) (Glycine acetyltransferase). | |||||
|
KBL_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 6) | NC score | 0.889807 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75600, Q96CA9 | Gene names | GCAT, KBL | |||
|
Domain Architecture |
|
|||||
| Description | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial precursor (EC 2.3.1.29) (AKB ligase) (Glycine acetyltransferase). | |||||
|
LCB2_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 7) | NC score | 0.813930 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97363, P97356 | Gene names | Sptlc2, Lcb2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 2 (EC 2.3.1.50) (Long chain base biosynthesis protein 2) (LCB 2) (Serine-palmitoyl-CoA transferase 2) (SPT 2). | |||||
|
LCB2_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 8) | NC score | 0.811708 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15270, Q16685 | Gene names | SPTLC2, KIAA0526, LCB2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 2 (EC 2.3.1.50) (Long chain base biosynthesis protein 2) (LCB 2) (Serine-palmitoyl-CoA transferase 2) (SPT 2). | |||||
|
LCB1_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 9) | NC score | 0.774564 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35704, O54813 | Gene names | Sptlc1, Lcb1 | |||
|
Domain Architecture |
|
|||||
| Description | Serine palmitoyltransferase 1 (EC 2.3.1.50) (Long chain base biosynthesis protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1). | |||||
|
LCB1_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 10) | NC score | 0.772738 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15269 | Gene names | SPTLC1, LCB1 | |||
|
Domain Architecture |
|
|||||
| Description | Serine palmitoyltransferase 1 (EC 2.3.1.50) (Long chain base biosynthesis protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1). | |||||
|
CCD37_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.047332 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q494V2, Q494V1, Q494V4, Q8N838 | Gene names | CCDC37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.041236 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.021428 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CEP57_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.025604 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.039543 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
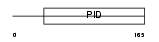
|
|||||
| Description | Disabled homolog 1. | |||||
|
NIT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.034959 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86X76, O76091 | Gene names | NIT1 | |||
|
Domain Architecture |
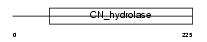
|
|||||
| Description | Nitrilase homolog 1 (EC 3.5.-.-). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.029869 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.024122 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ATE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.035472 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95260, O95261, Q8WW04 | Gene names | ATE1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.035621 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.023322 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.013613 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.006939 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
BMP2K_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.004332 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |
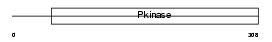
|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.021683 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
RCN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.021036 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
Domain Architecture |
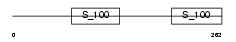
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.032062 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.023810 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
REXO4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.023478 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.024018 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
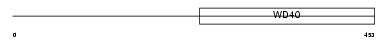
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.027210 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.022257 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.032170 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.028390 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
INGR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.022422 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15261 | Gene names | Ifngr1, Ifngr | |||
|
Domain Architecture |
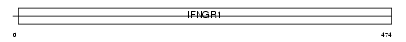
|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.015418 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
STK39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.000040 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
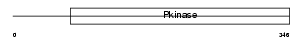
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.017075 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.005653 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RCN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.017983 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
Domain Architecture |
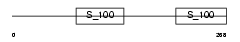
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.019302 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.012250 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.014905 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.018399 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.030891 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.014610 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
EPHB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | -0.000673 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.010600 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.021774 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.011867 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.010717 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.007920 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.014403 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
GLGB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.017185 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |
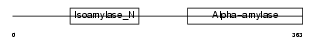
|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.003109 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
SAC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.014285 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |
|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.009167 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
ARP10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004888 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZB7, Q99LU1 | Gene names | Actr10, Act11, Actr11, Arp11 | |||
|
Domain Architecture |
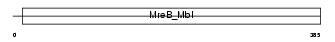
|
|||||
| Description | Actin-related protein 10. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.016531 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.016318 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
DNM1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.005135 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.012020 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.004041 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.013675 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SAC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.013329 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
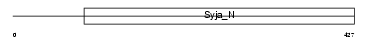
SYTL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.007972 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CT038_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.149226 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUV7, Q5T1U4, Q9H1L1, Q9H1Z0 | Gene names | C20orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf38. | |||||
|
HEM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VC19, Q64453 | Gene names | Alas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
HEM1_HUMAN
|
||||||
| NC score | 0.990798 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13196 | Gene names | ALAS1, ALASH | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
HEM0_MOUSE
|
||||||
| NC score | 0.984305 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08680, Q64452, Q9DCN0 | Gene names | Alas2 | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
HEM0_HUMAN
|
||||||
| NC score | 0.984095 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
KBL_MOUSE
|
||||||
| NC score | 0.893235 (rank : 5) | θ value | 8.28633e-54 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88986 | Gene names | Gcat, Kbl | |||
|
Domain Architecture |

|
|||||
| Description | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial precursor (EC 2.3.1.29) (AKB ligase) (Glycine acetyltransferase). | |||||
|
KBL_HUMAN
|
||||||
| NC score | 0.889807 (rank : 6) | θ value | 2.3352e-48 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75600, Q96CA9 | Gene names | GCAT, KBL | |||
|
Domain Architecture |

|
|||||
| Description | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial precursor (EC 2.3.1.29) (AKB ligase) (Glycine acetyltransferase). | |||||
|
LCB2_MOUSE
|
||||||
| NC score | 0.813930 (rank : 7) | θ value | 4.88048e-38 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97363, P97356 | Gene names | Sptlc2, Lcb2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 2 (EC 2.3.1.50) (Long chain base biosynthesis protein 2) (LCB 2) (Serine-palmitoyl-CoA transferase 2) (SPT 2). | |||||
|
LCB2_HUMAN
|
||||||
| NC score | 0.811708 (rank : 8) | θ value | 1.20211e-36 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15270, Q16685 | Gene names | SPTLC2, KIAA0526, LCB2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 2 (EC 2.3.1.50) (Long chain base biosynthesis protein 2) (LCB 2) (Serine-palmitoyl-CoA transferase 2) (SPT 2). | |||||
|
LCB1_MOUSE
|
||||||
| NC score | 0.774564 (rank : 9) | θ value | 4.01107e-24 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35704, O54813 | Gene names | Sptlc1, Lcb1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 1 (EC 2.3.1.50) (Long chain base biosynthesis protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1). | |||||
|
LCB1_HUMAN
|
||||||
| NC score | 0.772738 (rank : 10) | θ value | 8.93572e-24 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15269 | Gene names | SPTLC1, LCB1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine palmitoyltransferase 1 (EC 2.3.1.50) (Long chain base biosynthesis protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1). | |||||
|
CT038_HUMAN
|
||||||
| NC score | 0.149226 (rank : 11) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUV7, Q5T1U4, Q9H1L1, Q9H1Z0 | Gene names | C20orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf38. | |||||
|
CCD37_HUMAN
|
||||||
| NC score | 0.047332 (rank : 12) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q494V2, Q494V1, Q494V4, Q8N838 | Gene names | CCDC37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.041236 (rank : 13) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.039543 (rank : 14) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.035621 (rank : 15) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATE1_HUMAN
|
||||||
| NC score | 0.035472 (rank : 16) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95260, O95261, Q8WW04 | Gene names | ATE1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
NIT1_HUMAN
|
||||||
| NC score | 0.034959 (rank : 17) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86X76, O76091 | Gene names | NIT1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitrilase homolog 1 (EC 3.5.-.-). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.032170 (rank : 18) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.032062 (rank : 19) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.030891 (rank : 20) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.029869 (rank : 21) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.028390 (rank : 22) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.027210 (rank : 23) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CEP57_HUMAN
|
||||||
| NC score | 0.025604 (rank : 24) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.024122 (rank : 25) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.024018 (rank : 26) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.023810 (rank : 27) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
REXO4_HUMAN
|
||||||
| NC score | 0.023478 (rank : 28) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.023322 (rank : 29) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
INGR1_MOUSE
|
||||||
| NC score | 0.022422 (rank : 30) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15261 | Gene names | Ifngr1, Ifngr | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.022257 (rank : 31) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.021774 (rank : 32) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.021683 (rank : 33) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.021428 (rank : 34) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RCN1_MOUSE
|
||||||
| NC score | 0.021036 (rank : 35) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.019302 (rank : 36) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.018399 (rank : 37) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
RCN1_HUMAN
|
||||||
| NC score | 0.017983 (rank : 38) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
GLGB_HUMAN
|
||||||
| NC score | 0.017185 (rank : 39) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |

|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.017075 (rank : 40) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.016531 (rank : 41) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.016318 (rank : 42) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.015418 (rank : 43) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.014905 (rank : 44) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.014610 (rank : 45) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.014403 (rank : 46) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
SAC3_HUMAN
|
||||||
| NC score | 0.014285 (rank : 47) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |

|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.013675 (rank : 48) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.013613 (rank : 49) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.013329 (rank : 50) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.012250 (rank : 51) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.012020 (rank : 52) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.011867 (rank : 53) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.010717 (rank : 54) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.010600 (rank : 55) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.009167 (rank : 56) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.007972 (rank : 57) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.007920 (rank : 58) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.006939 (rank : 59) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.005653 (rank : 60) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
DNM1L_MOUSE
|
||||||
| NC score | 0.005135 (rank : 61) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
ARP10_MOUSE
|
||||||
| NC score | 0.004888 (rank : 62) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZB7, Q99LU1 | Gene names | Actr10, Act11, Actr11, Arp11 | |||
|
Domain Architecture |
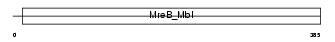
|
|||||
| Description | Actin-related protein 10. | |||||
|
BMP2K_HUMAN
|
||||||
| NC score | 0.004332 (rank : 63) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.004041 (rank : 64) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.003109 (rank : 65) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.000040 (rank : 66) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
EPHB3_MOUSE
|
||||||
| NC score | -0.000673 (rank : 67) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||