Please be patient as the page loads
|
HDGF_HUMAN
|
||||||
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HDGF_HUMAN
|
||||||
| θ value | 9.96626e-108 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 9.32813e-106 (rank : 2) | NC score | 0.963156 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
HDGR3_MOUSE
|
||||||
| θ value | 1.32293e-51 (rank : 3) | NC score | 0.956106 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 1.61718e-49 (rank : 4) | NC score | 0.948927 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 1.12253e-42 (rank : 5) | NC score | 0.820263 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 6) | NC score | 0.809602 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.046451 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.347747 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.094774 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.173891 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.170554 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
MSH6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.177498 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.211927 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.054154 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
DNM3B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.104382 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.203271 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
DNM3B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.105689 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.068094 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.055543 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.089164 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.044042 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.039669 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.083696 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.109940 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.038914 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.077609 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.096213 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.052969 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.147129 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.146551 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.089573 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.016780 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.037908 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.066825 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.033926 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.023441 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.068547 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.036060 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.020437 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.019429 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.051209 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SCG3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.041437 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.019613 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.022087 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.052243 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.023957 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ASPX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.044911 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.022754 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CU025_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.019812 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
NASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.060075 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.019183 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
RBM15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.026094 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
ERCC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.014230 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
GRWD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.005439 (rank : 99) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ67 | Gene names | GRWD1, GRWD, WDR28 | |||
|
Domain Architecture |
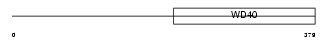
|
|||||
| Description | Glutamate-rich WD repeat-containing protein 1. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | -0.000165 (rank : 100) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.069194 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
USBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.016255 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
CASC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.028750 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
DEK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.054265 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.026181 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.014369 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
ARS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.062190 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ARS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.080628 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.084258 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.076423 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.052082 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.067738 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.079170 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.062686 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.073290 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.067096 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.073689 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.075210 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.073078 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
HMGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.088384 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05204, Q6FGI5, Q96C64 | Gene names | HMGN2, HMG17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-17 (High-mobility group nucleosome- binding domain-containing protein 2). | |||||
|
HMGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.069610 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09602 | Gene names | Hmgn2, Hmg-17, Hmg17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-17 (High-mobility group nucleosome- binding domain-containing protein 2). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.064767 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.073012 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.071562 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.057787 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054640 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.054527 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.061469 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.067991 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.067234 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.103600 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.075626 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.061712 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.058131 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051779 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.065060 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050341 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.060657 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.066678 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051432 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052370 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.065461 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.070762 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.050182 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050903 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
HDGF_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.96626e-108 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.963156 (rank : 2) | θ value | 9.32813e-106 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
HDGR3_MOUSE
|
||||||
| NC score | 0.956106 (rank : 3) | θ value | 1.32293e-51 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.948927 (rank : 4) | θ value | 1.61718e-49 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.820263 (rank : 5) | θ value | 1.12253e-42 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.809602 (rank : 6) | θ value | 8.04462e-41 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.347747 (rank : 7) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.211927 (rank : 8) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.203271 (rank : 9) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.177498 (rank : 10) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.173891 (rank : 11) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.170554 (rank : 12) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.147129 (rank : 13) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.146551 (rank : 14) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.109940 (rank : 15) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
DNM3B_HUMAN
|
||||||
| NC score | 0.105689 (rank : 16) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3B_MOUSE
|
||||||
| NC score | 0.104382 (rank : 17) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.103600 (rank : 18) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.096213 (rank : 19) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.094774 (rank : 20) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.089573 (rank : 21) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.089164 (rank : 22) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
HMGN2_HUMAN
|
||||||
| NC score | 0.088384 (rank : 23) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05204, Q6FGI5, Q96C64 | Gene names | HMGN2, HMG17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-17 (High-mobility group nucleosome- binding domain-containing protein 2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.084258 (rank : 24) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.083696 (rank : 25) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.080628 (rank : 26) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.079170 (rank : 27) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.077609 (rank : 28) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.076423 (rank : 29) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.075626 (rank : 30) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.075210 (rank : 31) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.073689 (rank : 32) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.073290 (rank : 33) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.073078 (rank : 34) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.073012 (rank : 35) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.071562 (rank : 36) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.070762 (rank : 37) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
HMGN2_MOUSE
|
||||||
| NC score | 0.069610 (rank : 38) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09602 | Gene names | Hmgn2, Hmg-17, Hmg17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-17 (High-mobility group nucleosome- binding domain-containing protein 2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.069194 (rank : 39) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.068547 (rank : 40) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.068094 (rank : 41) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.067991 (rank : 42) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.067738 (rank : 43) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.067234 (rank : 44) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.067096 (rank : 45) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.066825 (rank : 46) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.066678 (rank : 47) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.065461 (rank : 48) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.065060 (rank : 49) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.064767 (rank : 50) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.062686 (rank : 51) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.062190 (rank : 52) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.061712 (rank : 53) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.061469 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.060657 (rank : 55) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.060075 (rank : 56) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.058131 (rank : 57) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.057787 (rank : 58) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.055543 (rank : 59) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.054640 (rank : 60) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.054527 (rank : 61) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.054265 (rank : 62) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.054154 (rank : 63) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.052969 (rank : 64) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.052370 (rank : 65) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.052243 (rank : 66) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.052082 (rank : 67) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.051779 (rank : 68) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
TCAL2_HUMAN
|
||||||
| NC score | 0.051432 (rank : 69) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.051209 (rank : 70) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.050903 (rank : 71) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.050341 (rank : 72) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.050182 (rank : 73) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.046451 (rank : 74) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.044911 (rank : 75) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.044042 (rank : 76) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SCG3_MOUSE
|
||||||
| NC score | 0.041437 (rank : 77) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.039669 (rank : 78) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.038914 (rank : 79) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.037908 (rank : 80) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.036060 (rank : 81) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.033926 (rank : 82) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
CASC3_HUMAN
|
||||||
| NC score | 0.028750 (rank : 83) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15234 | Gene names | CASC3, MLN51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein) (Metastatic lymph node protein 51) (Protein MLN 51) (Protein barentsz) (Btz). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.026181 (rank : 84) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.026094 (rank : 85) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.023957 (rank : 86) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.023441 (rank : 87) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.022754 (rank : 88) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.022087 (rank : 89) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.020437 (rank : 90) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CU025_HUMAN
|
||||||
| NC score | 0.019812 (rank : 91) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.019613 (rank : 92) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.019429 (rank : 93) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.019183 (rank : 94) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.016780 (rank : 95) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
USBP1_MOUSE
|
||||||
| NC score | 0.016255 (rank : 96) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.014369 (rank : 97) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
ERCC4_MOUSE
|
||||||
| NC score | 0.014230 (rank : 98) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
GRWD1_HUMAN
|
||||||
| NC score | 0.005439 (rank : 99) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ67 | Gene names | GRWD1, GRWD, WDR28 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate-rich WD repeat-containing protein 1. | |||||
|
LATS1_MOUSE
|
||||||
| NC score | -0.000165 (rank : 100) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||