Please be patient as the page loads
|
CJ095_HUMAN
|
||||||
| SwissProt Accessions | Q9H7T3 | Gene names | C10orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf95. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CJ095_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H7T3 | Gene names | C10orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf95. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 2) | NC score | 0.124501 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN598_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 3) | NC score | 0.131820 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YR4, Q6KAT0, Q8R3S1 | Gene names | Znf598, Zfp598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.057907 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.038313 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
EPHB4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.023400 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.070401 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.094154 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ATS16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.043408 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE57, Q8IVE2 | Gene names | ADAMTS16, KIAA2029 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-16 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 16) (ADAM-TS 16) (ADAM-TS16). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.070533 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
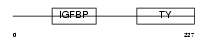
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
TSP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.060789 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.024322 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.083094 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.063603 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.035444 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.063794 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.043644 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.045469 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.034372 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.047933 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GFRA4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.071181 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJT2, Q9JJT3, Q9JJT4, Q9JJT6, Q9JJT7, Q9JJT8 | Gene names | Gfra4 | |||
|
Domain Architecture |
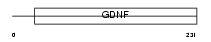
|
|||||
| Description | GDNF family receptor alpha-4 precursor (GFR-alpha-4) (Persephin receptor). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.049351 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
KR103_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.039137 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.021114 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.013431 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.041639 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
GFRA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.069396 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60609, Q6UW20, Q8IUZ2 | Gene names | GFRA3 | |||
|
Domain Architecture |
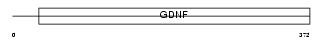
|
|||||
| Description | GDNF family receptor alpha-3 precursor (GFR-alpha-3). | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.067279 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.026885 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.037720 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.060821 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.080637 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.051347 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
ERBB4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.019355 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
GFRA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.063578 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35118, O35325, O55243, Q6NZC2, Q8C8L9 | Gene names | Gfra3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GDNF family receptor alpha-3 precursor (GFR-alpha-3). | |||||
|
HXD12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.012037 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.049885 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TSP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.054453 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.044310 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PROP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.044011 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.019692 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.010835 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.028786 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.042648 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
ES8L1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.016279 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.019004 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.031362 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.018808 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
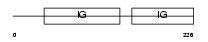
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.014772 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.017963 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.052689 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.059945 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.052760 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.067876 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.051005 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.050366 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052355 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.060027 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.063215 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.062870 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053663 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.058705 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.058727 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.054079 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.087360 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.064200 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.056073 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.082637 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052209 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
NACA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052257 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.060433 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050511 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055312 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051222 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.058412 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.058574 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.085621 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.079149 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.068270 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.067324 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.064364 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.050838 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050184 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.090567 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.083877 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.079095 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.072702 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.060313 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.062727 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.102629 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.056896 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.060157 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.064086 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.059873 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.098183 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CJ095_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.10721e-57 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H7T3 | Gene names | C10orf95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf95. | |||||
|
ZN598_MOUSE
|
||||||
| NC score | 0.131820 (rank : 2) | θ value | 0.0563607 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YR4, Q6KAT0, Q8R3S1 | Gene names | Znf598, Zfp598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.124501 (rank : 3) | θ value | 0.000158464 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.102629 (rank : 4) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.098183 (rank : 5) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.094154 (rank : 6) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.090567 (rank : 7) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.087360 (rank : 8) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.085621 (rank : 9) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.083877 (rank : 10) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.083094 (rank : 11) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.082637 (rank : 12) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.080637 (rank : 13) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.079149 (rank : 14) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.079095 (rank : 15) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.072702 (rank : 16) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GFRA4_MOUSE
|
||||||
| NC score | 0.071181 (rank : 17) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJT2, Q9JJT3, Q9JJT4, Q9JJT6, Q9JJT7, Q9JJT8 | Gene names | Gfra4 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF family receptor alpha-4 precursor (GFR-alpha-4) (Persephin receptor). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.070533 (rank : 18) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.070401 (rank : 19) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
GFRA3_HUMAN
|
||||||
| NC score | 0.069396 (rank : 20) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60609, Q6UW20, Q8IUZ2 | Gene names | GFRA3 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF family receptor alpha-3 precursor (GFR-alpha-3). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.068270 (rank : 21) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.067876 (rank : 22) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.067324 (rank : 23) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.067279 (rank : 24) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.064364 (rank : 25) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.064200 (rank : 26) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.064086 (rank : 27) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.063794 (rank : 28) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.063603 (rank : 29) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GFRA3_MOUSE
|
||||||
| NC score | 0.063578 (rank : 30) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35118, O35325, O55243, Q6NZC2, Q8C8L9 | Gene names | Gfra3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GDNF family receptor alpha-3 precursor (GFR-alpha-3). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.063215 (rank : 31) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.062870 (rank : 32) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.062727 (rank : 33) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.060821 (rank : 34) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
TSP2_MOUSE
|
||||||
| NC score | 0.060789 (rank : 35) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.060433 (rank : 36) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.060313 (rank : 37) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.060157 (rank : 38) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.060027 (rank : 39) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.059945 (rank : 40) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.059873 (rank : 41) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.058727 (rank : 42) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.058705 (rank : 43) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.058574 (rank : 44) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.058412 (rank : 45) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.057907 (rank : 46) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.056896 (rank : 47) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.056073 (rank : 48) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.055312 (rank : 49) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TSP2_HUMAN
|
||||||
| NC score | 0.054453 (rank : 50) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.054079 (rank : 51) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.053663 (rank : 52) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.052760 (rank : 53) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.052689 (rank : 54) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.052355 (rank : 55) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NACA_MOUSE
|
||||||
| NC score | 0.052257 (rank : 56) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
NACA_HUMAN
|
||||||
| NC score | 0.052209 (rank : 57) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.051347 (rank : 58) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.051222 (rank : 59) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.051005 (rank : 60) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.050838 (rank : 61) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.050511 (rank : 62) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.050366 (rank : 63) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.050184 (rank : 64) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.049885 (rank : 65) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.049351 (rank : 66) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.047933 (rank : 67) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.045469 (rank : 68) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.044310 (rank : 69) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.044011 (rank : 70) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.043644 (rank : 71) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
ATS16_HUMAN
|
||||||
| NC score | 0.043408 (rank : 72) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE57, Q8IVE2 | Gene names | ADAMTS16, KIAA2029 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-16 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 16) (ADAM-TS 16) (ADAM-TS16). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.042648 (rank : 73) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.041639 (rank : 74) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.039137 (rank : 75) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.038313 (rank : 76) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.037720 (rank : 77) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.035444 (rank : 78) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.034372 (rank : 79) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.031362 (rank : 80) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.028786 (rank : 81) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.026885 (rank : 82) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.024322 (rank : 83) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
EPHB4_HUMAN
|
||||||
| NC score | 0.023400 (rank : 84) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.021114 (rank : 85) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.019692 (rank : 86) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
ERBB4_HUMAN
|
||||||
| NC score | 0.019355 (rank : 87) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.019004 (rank : 88) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.018808 (rank : 89) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.017963 (rank : 90) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
ES8L1_MOUSE
|
||||||
| NC score | 0.016279 (rank : 91) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.014772 (rank : 92) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.013431 (rank : 93) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
HXD12_MOUSE
|
||||||
| NC score | 0.012037 (rank : 94) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23812 | Gene names | Hoxd12, Hox-4.7, Hoxd-12 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4.7) (Hox-5.6). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.010835 (rank : 95) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||