Please be patient as the page loads
|
XLKD1_HUMAN
|
||||||
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
XLKD1_HUMAN
|
||||||
| θ value | 1.98498e-148 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| θ value | 2.07809e-105 (rank : 2) | NC score | 0.987280 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
CD44_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 3) | NC score | 0.608778 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |
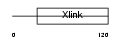
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
CD44_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 4) | NC score | 0.601322 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |
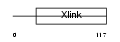
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 5) | NC score | 0.433620 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 6) | NC score | 0.434181 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.289805 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 8) | NC score | 0.377432 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 9) | NC score | 0.283716 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
HPLN3_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 10) | NC score | 0.494941 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |
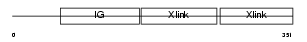
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.253822 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 12) | NC score | 0.331011 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 13) | NC score | 0.255047 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 14) | NC score | 0.315576 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
HPLN3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 15) | NC score | 0.487508 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |
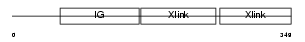
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 16) | NC score | 0.311043 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 17) | NC score | 0.354121 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
HPLN2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.480088 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |
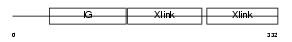
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 19) | NC score | 0.357775 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 20) | NC score | 0.302545 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
HPLN2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 21) | NC score | 0.479550 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |
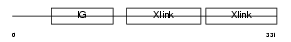
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 22) | NC score | 0.473348 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |
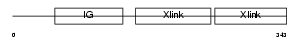
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 23) | NC score | 0.473729 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |
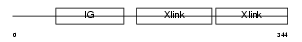
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN4_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 24) | NC score | 0.463396 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |
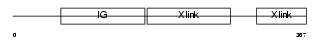
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 25) | NC score | 0.299455 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.466934 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
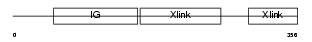
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.018250 (rank : 147) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.029331 (rank : 146) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.010215 (rank : 148) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.008129 (rank : 150) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.010150 (rank : 149) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
ASGR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.081089 (rank : 43) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |
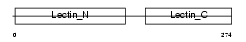
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
ASGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.079541 (rank : 47) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |
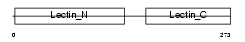
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
ASGR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.076486 (rank : 55) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |
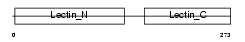
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.079281 (rank : 48) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
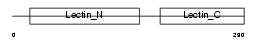
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.050666 (rank : 143) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
BGH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.065048 (rank : 84) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15582, O14471, O14472, O14476, O43216, O43217, O43218, O43219 | Gene names | TGFBI, BIGH3 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3) (Kerato-epithelin) (RGD-containing collagen-associated protein) (RGD-CAP). | |||||
|
BGH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.063097 (rank : 90) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P82198, Q3U9R1 | Gene names | Tgfbi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3). | |||||
|
C209A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.070344 (rank : 68) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |
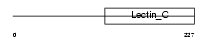
|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
C209B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.066685 (rank : 78) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
C209C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.080171 (rank : 45) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |
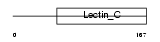
|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
C209D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.068318 (rank : 72) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
C209E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.078264 (rank : 51) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
CD209_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.067399 (rank : 76) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
CD302_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.068760 (rank : 71) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.072032 (rank : 62) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CD69_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.065002 (rank : 85) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |
|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CD69_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.053862 (rank : 127) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.080511 (rank : 44) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.071578 (rank : 65) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CLC10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.081664 (rank : 40) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
CLC3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.065220 (rank : 83) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |
|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.066474 (rank : 79) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.070286 (rank : 69) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
CLC4A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.064956 (rank : 86) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
CLC4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.062750 (rank : 93) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
CLC4D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.092985 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
CLC4D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.089384 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
CLC4E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.073409 (rank : 60) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.078644 (rank : 50) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.064907 (rank : 87) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
CLC4G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.083658 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.083147 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.077196 (rank : 53) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.076747 (rank : 54) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4M_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.061847 (rank : 96) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
CLC6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.058230 (rank : 113) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.059688 (rank : 103) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.053166 (rank : 128) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.053116 (rank : 129) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.054678 (rank : 124) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.071982 (rank : 64) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.070966 (rank : 67) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CXAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051849 (rank : 134) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |
|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.060590 (rank : 101) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.059314 (rank : 104) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.056341 (rank : 120) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.055733 (rank : 122) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.056727 (rank : 118) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.059243 (rank : 106) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055959 (rank : 121) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057217 (rank : 116) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057358 (rank : 115) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DNER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059028 (rank : 108) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051963 (rank : 133) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
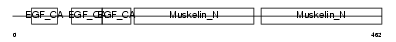
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050938 (rank : 140) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
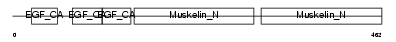
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.061448 (rank : 99) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.062413 (rank : 95) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
FCER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.089827 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
FCER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.081566 (rank : 41) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
GPA33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.056760 (rank : 117) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99795 | Gene names | GPA33 | |||
|
Domain Architecture |
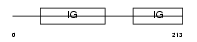
|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051135 (rank : 139) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051215 (rank : 138) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
KLRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.050800 (rank : 142) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |
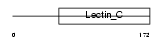
|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
KLRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.064870 (rank : 88) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.066098 (rank : 81) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052521 (rank : 131) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.058597 (rank : 109) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059243 (rank : 105) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.056452 (rank : 119) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.054423 (rank : 125) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058377 (rank : 112) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.065841 (rank : 82) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.050279 (rank : 145) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.075952 (rank : 58) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051707 (rank : 135) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.067510 (rank : 75) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |
|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
LIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.068246 (rank : 73) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |
|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
LORI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.073253 (rank : 61) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.141197 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
LY75_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.062880 (rank : 92) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
LY75_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.062932 (rank : 91) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
LYAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051573 (rank : 136) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050938 (rank : 141) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
LYAM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.058568 (rank : 110) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
MBL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.073827 (rank : 59) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |
|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
MBL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.061468 (rank : 98) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
MBL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.071985 (rank : 63) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.079630 (rank : 46) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.071368 (rank : 66) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.069252 (rank : 70) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.082052 (rank : 39) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.082955 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
NID2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051476 (rank : 137) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NKG2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.061767 (rank : 97) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |
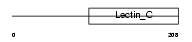
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
NKG2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.067867 (rank : 74) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |
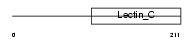
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
PHXR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.079122 (rank : 49) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
POSTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053950 (rank : 126) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15063, Q15064, Q5VSY5, Q8IZF9 | Gene names | POSTN, OSF2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
POSTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054684 (rank : 123) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62009, Q8BMJ6, Q8K1K0 | Gene names | Postn, Osf2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.063295 (rank : 89) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
PRG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.062544 (rank : 94) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
REG1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.077570 (rank : 52) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |
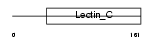
|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.067208 (rank : 77) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |
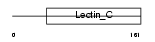
|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
REG3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.076439 (rank : 56) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |
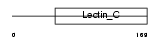
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
REG3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.076434 (rank : 57) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |
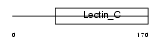
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
REG3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.083024 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |
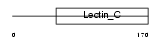
|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
REG3G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.066214 (rank : 80) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
REG3G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.083564 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |
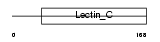
|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
REG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.091695 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
REG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.099209 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
SFTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.059149 (rank : 107) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
SFTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.058390 (rank : 111) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
SFTPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.052721 (rank : 130) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35242 | Gene names | Sftpa1, Sftp-1, Sftp1, Sftpa | |||
|
Domain Architecture |
|
|||||
| Description | Pulmonary surfactant-associated protein A precursor (SP-A) (PSP-A) (PSAP). | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.057408 (rank : 114) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
TETN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.087024 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
TETN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.081564 (rank : 42) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
UROM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.051979 (rank : 132) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
UROM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.050660 (rank : 144) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.061368 (rank : 100) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.060308 (rank : 102) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
XLKD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.98498e-148 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| NC score | 0.987280 (rank : 2) | θ value | 2.07809e-105 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
CD44_MOUSE
|
||||||
| NC score | 0.608778 (rank : 3) | θ value | 8.95645e-16 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
CD44_HUMAN
|
||||||
| NC score | 0.601322 (rank : 4) | θ value | 6.41864e-14 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
HPLN3_HUMAN
|
||||||
| NC score | 0.494941 (rank : 5) | θ value | 6.43352e-06 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
HPLN3_MOUSE
|
||||||
| NC score | 0.487508 (rank : 6) | θ value | 0.000158464 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
HPLN2_MOUSE
|
||||||
| NC score | 0.480088 (rank : 7) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_HUMAN
|
||||||
| NC score | 0.479550 (rank : 8) | θ value | 0.000786445 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN1_MOUSE
|
||||||
| NC score | 0.473729 (rank : 9) | θ value | 0.00134147 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_HUMAN
|
||||||
| NC score | 0.473348 (rank : 10) | θ value | 0.00102713 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.466934 (rank : 11) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
HPLN4_HUMAN
|
||||||
| NC score | 0.463396 (rank : 12) | θ value | 0.00175202 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.434181 (rank : 13) | θ value | 1.06045e-08 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.433620 (rank : 14) | θ value | 8.11959e-09 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.377432 (rank : 15) | θ value | 2.88788e-06 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.357775 (rank : 16) | θ value | 0.000602161 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.354121 (rank : 17) | θ value | 0.000461057 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.331011 (rank : 18) | θ value | 5.44631e-05 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.315576 (rank : 19) | θ value | 7.1131e-05 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.311043 (rank : 20) | θ value | 0.000270298 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.302545 (rank : 21) | θ value | 0.000786445 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.299455 (rank : 22) | θ value | 0.00228821 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.289805 (rank : 23) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.283716 (rank : 24) | θ value | 2.88788e-06 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.255047 (rank : 25) | θ value | 5.44631e-05 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.253822 (rank : 26) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.141197 (rank : 27) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
REG4_MOUSE
|
||||||
| NC score | 0.099209 (rank : 28) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
CLC4D_HUMAN
|
||||||
| NC score | 0.092985 (rank : 29) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
REG4_HUMAN
|
||||||
| NC score | 0.091695 (rank : 30) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
FCER2_HUMAN
|
||||||
| NC score | 0.089827 (rank : 31) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
CLC4D_MOUSE
|
||||||
| NC score | 0.089384 (rank : 32) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
TETN_HUMAN
|
||||||
| NC score | 0.087024 (rank : 33) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
CLC4G_HUMAN
|
||||||
| NC score | 0.083658 (rank : 34) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
REG3G_MOUSE
|
||||||
| NC score | 0.083564 (rank : 35) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
CLC4G_MOUSE
|
||||||
| NC score | 0.083147 (rank : 36) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
REG3B_MOUSE
|
||||||
| NC score | 0.083024 (rank : 37) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.082955 (rank : 38) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.082052 (rank : 39) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.081664 (rank : 40) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
FCER2_MOUSE
|
||||||
| NC score | 0.081566 (rank : 41) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
TETN_MOUSE
|
||||||
| NC score | 0.081564 (rank : 42) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
ASGR1_HUMAN
|
||||||
| NC score | 0.081089 (rank : 43) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07306 | Gene names | ASGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin H1). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.080511 (rank : 44) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
C209C_MOUSE
|
||||||
| NC score | 0.080171 (rank : 45) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.079630 (rank : 46) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
ASGR1_MOUSE
|
||||||
| NC score | 0.079541 (rank : 47) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.079281 (rank : 48) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
PHXR5_MOUSE
|
||||||
| NC score | 0.079122 (rank : 49) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08399 | Gene names | Phxr5, Per | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Per-hexamer repeat protein 5. | |||||
|
CLC4E_MOUSE
|
||||||
| NC score | 0.078644 (rank : 50) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
C209E_MOUSE
|
||||||
| NC score | 0.078264 (rank : 51) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
REG1A_HUMAN
|
||||||
| NC score | 0.077570 (rank : 52) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
CLC4K_HUMAN
|
||||||
| NC score | 0.077196 (rank : 53) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| NC score | 0.076747 (rank : 54) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
ASGR2_HUMAN
|
||||||
| NC score | 0.076486 (rank : 55) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
REG3A_HUMAN
|
||||||
| NC score | 0.076439 (rank : 56) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
REG3A_MOUSE
|
||||||
| NC score | 0.076434 (rank : 57) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.075952 (rank : 58) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
MBL1_MOUSE
|
||||||
| NC score | 0.073827 (rank : 59) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
CLC4E_HUMAN
|
||||||
| NC score | 0.073409 (rank : 60) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
LORI_HUMAN
|
||||||
| NC score | 0.073253 (rank : 61) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23490 | Gene names | LOR, LRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.072032 (rank : 62) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
MBL2_MOUSE
|
||||||
| NC score | 0.071985 (rank : 63) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.071982 (rank : 64) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.071578 (rank : 65) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.071368 (rank : 66) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.070966 (rank : 67) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
C209A_MOUSE
|
||||||
| NC score | 0.070344 (rank : 68) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
CLC4A_HUMAN
|
||||||
| NC score | 0.070286 (rank : 69) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.069252 (rank : 70) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.068760 (rank : 71) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
C209D_MOUSE
|
||||||
| NC score | 0.068318 (rank : 72) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
LIT2_MOUSE
|
||||||
| NC score | 0.068246 (rank : 73) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
NKG2D_MOUSE
|
||||||
| NC score | 0.067867 (rank : 74) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
LIT1_MOUSE
|
||||||
| NC score | 0.067510 (rank : 75) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
CD209_HUMAN
|
||||||
| NC score | 0.067399 (rank : 76) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
REG1B_HUMAN
|
||||||
| NC score | 0.067208 (rank : 77) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
C209B_MOUSE
|
||||||
| NC score | 0.066685 (rank : 78) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
CLC3A_MOUSE
|
||||||
| NC score | 0.066474 (rank : 79) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
REG3G_HUMAN
|
||||||
| NC score | 0.066214 (rank : 80) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.066098 (rank : 81) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.065841 (rank : 82) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
CLC3A_HUMAN
|
||||||
| NC score | 0.065220 (rank : 83) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
BGH3_HUMAN
|
||||||
| NC score | 0.065048 (rank : 84) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15582, O14471, O14472, O14476, O43216, O43217, O43218, O43219 | Gene names | TGFBI, BIGH3 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3) (Kerato-epithelin) (RGD-containing collagen-associated protein) (RGD-CAP). | |||||
|
CD69_HUMAN
|
||||||
| NC score | 0.065002 (rank : 85) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |

|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CLC4A_MOUSE
|
||||||
| NC score | 0.064956 (rank : 86) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.064907 (rank : 87) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
KLRF1_HUMAN
|
||||||
| NC score | 0.064870 (rank : 88) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.063295 (rank : 89) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
BGH3_MOUSE
|
||||||
| NC score | 0.063097 (rank : 90) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P82198, Q3U9R1 | Gene names | Tgfbi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.062932 (rank : 91) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.062880 (rank : 92) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
CLC4C_HUMAN
|
||||||
| NC score | 0.062750 (rank : 93) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.062544 (rank : 94) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.062413 (rank : 95) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
CLC4M_HUMAN
|
||||||
| NC score | 0.061847 (rank : 96) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
NKG2D_HUMAN
|
||||||
| NC score | 0.061767 (rank : 97) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
MBL2_HUMAN
|
||||||
| NC score | 0.061468 (rank : 98) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.061448 (rank : 99) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.061368 (rank : 100) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.060590 (rank : 101) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.060308 (rank : 102) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
CLC6A_MOUSE
|
||||||
| NC score | 0.059688 (rank : 103) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.059314 (rank : 104) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.059243 (rank : 105) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.059243 (rank : 106) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
SFTA1_HUMAN
|
||||||
| NC score | 0.059149 (rank : 107) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
DNER_MOUSE
|
||||||
| NC score | 0.059028 (rank : 108) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.058597 (rank : 109) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
LYAM2_MOUSE
|
||||||
| NC score | 0.058568 (rank : 110) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
SFTA2_HUMAN
|
||||||
| NC score | 0.058390 (rank : 111) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.058377 (rank : 112) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
CLC6A_HUMAN
|
||||||
| NC score | 0.058230 (rank : 113) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.057408 (rank : 114) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.057358 (rank : 115) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.057217 (rank : 116) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
GPA33_HUMAN
|
||||||
| NC score | 0.056760 (rank : 117) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99795 | Gene names | GPA33 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.056727 (rank : 118) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.056452 (rank : 119) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.056341 (rank : 120) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.055959 (rank : 121) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.055733 (rank : 122) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
POSTN_MOUSE
|
||||||
| NC score | 0.054684 (rank : 123) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62009, Q8BMJ6, Q8K1K0 | Gene names | Postn, Osf2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.054678 (rank : 124) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.054423 (rank : 125) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
POSTN_HUMAN
|
||||||
| NC score | 0.053950 (rank : 126) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15063, Q15064, Q5VSY5, Q8IZF9 | Gene names | POSTN, OSF2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
CD69_MOUSE
|
||||||
| NC score | 0.053862 (rank : 127) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.053166 (rank : 128) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.053116 (rank : 129) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
SFTPA_MOUSE
|
||||||
| NC score | 0.052721 (rank : 130) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35242 | Gene names | Sftpa1, Sftp-1, Sftp1, Sftpa | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein A precursor (SP-A) (PSP-A) (PSAP). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.052521 (rank : 131) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
UROM_HUMAN
|
||||||
| NC score | 0.051979 (rank : 132) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.051963 (rank : 133) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
CXAR_HUMAN
|
||||||
| NC score | 0.051849 (rank : 134) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |

|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.051707 (rank : 135) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.051573 (rank : 136) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.051476 (rank : 137) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.051215 (rank : 138) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.051135 (rank : 139) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.050938 (rank : 140) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
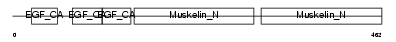
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
LYAM2_HUMAN
|
||||||
| NC score | 0.050938 (rank : 141) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
KLRD1_HUMAN
|
||||||
| NC score | 0.050800 (rank : 142) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.050666 (rank : 143) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
UROM_MOUSE
|
||||||
| NC score | 0.050660 (rank : 144) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.050279 (rank : 145) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.029331 (rank : 146) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.018250 (rank : 147) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.010215 (rank : 148) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.010150 (rank : 149) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.008129 (rank : 150) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||