Please be patient as the page loads
|
UQCR2_MOUSE
|
||||||
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
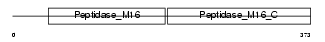
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UQCR2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998540 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
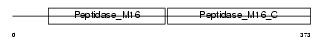
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 3) | NC score | 0.801733 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
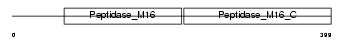
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 4) | NC score | 0.793715 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
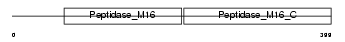
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 5) | NC score | 0.805654 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 6) | NC score | 0.799593 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 7) | NC score | 0.769531 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 8) | NC score | 0.754880 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
PDK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.022770 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFP9 | Gene names | Pdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 1). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.011784 (rank : 17) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
PDK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.021168 (rank : 14) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15118 | Gene names | PDK1 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 1). | |||||
|
HNRPM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.016575 (rank : 15) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.015742 (rank : 16) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
IDE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.079138 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.084719 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.091385 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.087604 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.998540 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.805654 (rank : 3) | θ value | 3.74554e-30 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.801733 (rank : 4) | θ value | 1.46948e-34 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.799593 (rank : 5) | θ value | 9.2256e-29 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.793715 (rank : 6) | θ value | 1.98606e-31 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.769531 (rank : 7) | θ value | 2.59989e-23 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.754880 (rank : 8) | θ value | 4.58923e-20 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.091385 (rank : 9) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.087604 (rank : 10) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.084719 (rank : 11) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.079138 (rank : 12) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
PDK1_MOUSE
|
||||||
| NC score | 0.022770 (rank : 13) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFP9 | Gene names | Pdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 1). | |||||
|
PDK1_HUMAN
|
||||||
| NC score | 0.021168 (rank : 14) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15118 | Gene names | PDK1 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial precursor (EC 2.7.11.2) (Pyruvate dehydrogenase kinase isoform 1). | |||||
|
HNRPM_MOUSE
|
||||||
| NC score | 0.016575 (rank : 15) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_HUMAN
|
||||||
| NC score | 0.015742 (rank : 16) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.011784 (rank : 17) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||