Please be patient as the page loads
|
UQCR1_MOUSE
|
||||||
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
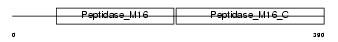
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UQCR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997726 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
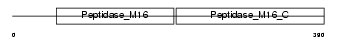
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 8.32034e-155 (rank : 3) | NC score | 0.988443 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
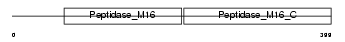
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 2.67656e-153 (rank : 4) | NC score | 0.988201 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
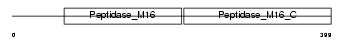
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 5) | NC score | 0.863995 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
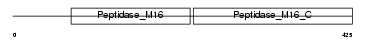
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 6) | NC score | 0.864338 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
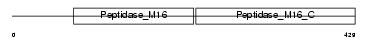
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 7) | NC score | 0.769531 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
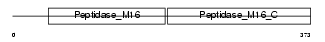
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 8) | NC score | 0.762831 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
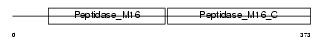
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 9) | NC score | 0.246734 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.239931 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.230003 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.221307 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
K0240_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.028072 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
TYDP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.025156 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.019263 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.018804 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.013873 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
K0240_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.021783 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
SYT14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.005469 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.997726 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.988443 (rank : 3) | θ value | 8.32034e-155 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.988201 (rank : 4) | θ value | 2.67656e-153 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.864338 (rank : 5) | θ value | 6.58091e-43 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.863995 (rank : 6) | θ value | 1.73182e-43 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.769531 (rank : 7) | θ value | 2.59989e-23 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.762831 (rank : 8) | θ value | 4.43474e-23 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.246734 (rank : 9) | θ value | 0.000270298 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.239931 (rank : 10) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.230003 (rank : 11) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.221307 (rank : 12) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.028072 (rank : 13) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
TYDP1_MOUSE
|
||||||
| NC score | 0.025156 (rank : 14) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.021783 (rank : 15) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.019263 (rank : 16) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.018804 (rank : 17) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.013873 (rank : 18) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SYT14_HUMAN
|
||||||
| NC score | 0.005469 (rank : 19) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||