Please be patient as the page loads
|
MPPB_MOUSE
|
||||||
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
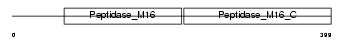
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MPPB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999379 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
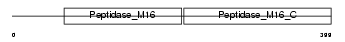
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_HUMAN
|
||||||
| θ value | 2.04937e-153 (rank : 3) | NC score | 0.986846 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
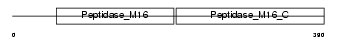
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 2.67656e-153 (rank : 4) | NC score | 0.988201 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
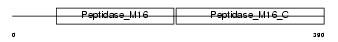
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 1.04822e-48 (rank : 5) | NC score | 0.876117 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
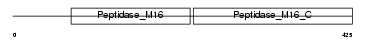
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 6) | NC score | 0.876799 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
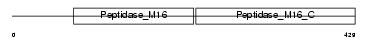
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 7) | NC score | 0.801733 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
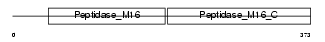
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 8) | NC score | 0.792676 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
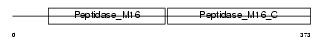
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 9) | NC score | 0.246865 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 10) | NC score | 0.239505 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 11) | NC score | 0.234758 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 12) | NC score | 0.226071 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
KIFC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.015093 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.016379 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
KIFC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.012355 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.005527 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CCD42_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.007555 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.999379 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.988201 (rank : 3) | θ value | 2.67656e-153 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.986846 (rank : 4) | θ value | 2.04937e-153 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.876799 (rank : 5) | θ value | 2.3352e-48 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.876117 (rank : 6) | θ value | 1.04822e-48 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.801733 (rank : 7) | θ value | 1.46948e-34 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.792676 (rank : 8) | θ value | 4.14116e-29 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.246865 (rank : 9) | θ value | 0.000270298 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.239505 (rank : 10) | θ value | 0.000786445 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.234758 (rank : 11) | θ value | 0.00228821 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.226071 (rank : 12) | θ value | 0.00665767 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.016379 (rank : 13) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
KIFC3_MOUSE
|
||||||
| NC score | 0.015093 (rank : 14) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
KIFC3_HUMAN
|
||||||
| NC score | 0.012355 (rank : 15) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
CCD42_HUMAN
|
||||||
| NC score | 0.007555 (rank : 16) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.005527 (rank : 17) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||