Please be patient as the page loads
|
UQCR1_HUMAN
|
||||||
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UQCR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997726 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| θ value | 8.32034e-155 (rank : 3) | NC score | 0.987826 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| θ value | 2.04937e-153 (rank : 4) | NC score | 0.986846 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 5) | NC score | 0.858043 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |
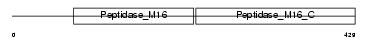
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 6) | NC score | 0.858001 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |
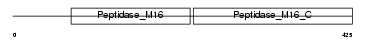
|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 7) | NC score | 0.748035 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |
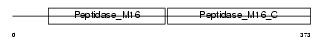
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 8) | NC score | 0.754880 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |
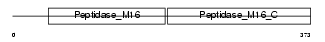
|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
IDE_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 9) | NC score | 0.253612 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 10) | NC score | 0.261086 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.254158 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.244784 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.007974 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.003830 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.009381 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.003425 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ACHA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.002622 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.007689 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
UQCR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31930, Q96DD2 | Gene names | UQCRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
UQCR1_MOUSE
|
||||||
| NC score | 0.997726 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZ13, Q9CWL6 | Gene names | Uqcrc1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2). | |||||
|
MPPB_HUMAN
|
||||||
| NC score | 0.987826 (rank : 3) | θ value | 8.32034e-155 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75439, O60416, Q96FV4 | Gene names | PMPCB, MPPB | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPB_MOUSE
|
||||||
| NC score | 0.986846 (rank : 4) | θ value | 2.04937e-153 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CXT8 | Gene names | Pmpcb | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase subunit beta, mitochondrial precursor (EC 3.4.24.64) (Beta-MPP) (P-52). | |||||
|
MPPA_HUMAN
|
||||||
| NC score | 0.858043 (rank : 5) | θ value | 8.04462e-41 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q10713, Q16639, Q8N513 | Gene names | PMPCA, KIAA0123, MPPA | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
MPPA_MOUSE
|
||||||
| NC score | 0.858001 (rank : 6) | θ value | 1.79215e-40 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC61, Q3TF19 | Gene names | Pmpca, Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial-processing peptidase alpha subunit, mitochondrial precursor (EC 3.4.24.64) (Alpha-MPP) (P-55). | |||||
|
UQCR2_MOUSE
|
||||||
| NC score | 0.754880 (rank : 7) | θ value | 4.58923e-20 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB77 | Gene names | Uqcrc2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
UQCR2_HUMAN
|
||||||
| NC score | 0.748035 (rank : 8) | θ value | 3.51386e-20 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22695, Q9BQ05 | Gene names | UQCRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinol-cytochrome-c reductase complex core protein 2, mitochondrial precursor (EC 1.10.2.2) (Complex III subunit II). | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.261086 (rank : 9) | θ value | 0.000461057 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.254158 (rank : 10) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
IDE_MOUSE
|
||||||
| NC score | 0.253612 (rank : 11) | θ value | 0.000121331 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHR7 | Gene names | Ide | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
IDE_HUMAN
|
||||||
| NC score | 0.244784 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14735 | Gene names | IDE | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-degrading enzyme (EC 3.4.24.56) (Insulysin) (Insulinase) (Insulin protease). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.009381 (rank : 13) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.007974 (rank : 14) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.007689 (rank : 15) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.003830 (rank : 16) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.003425 (rank : 17) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ACHA2_HUMAN
|
||||||
| NC score | 0.002622 (rank : 18) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||