Please be patient as the page loads
|
TPH1_HUMAN
|
||||||
| SwissProt Accessions | P17752, O95188, O95189, Q16736 | Gene names | TPH1, TPH, TRPH | |||
|
Domain Architecture |
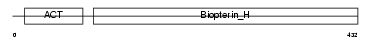
|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TPH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17752, O95188, O95189, Q16736 | Gene names | TPH1, TPH, TRPH | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
TPH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999098 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17532 | Gene names | Tph1, Tph | |||
|
Domain Architecture |
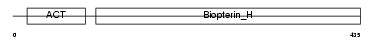
|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
TPH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996531 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWU9 | Gene names | TPH2, NTPH | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 2 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 2) (Neuronal tryptophan hydroxylase). | |||||
|
TPH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996424 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGV2 | Gene names | Tph2, Ntph | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 2 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 2) (Neuronal tryptophan hydroxylase). | |||||
|
PH4H_MOUSE
|
||||||
| θ value | 5.78879e-140 (rank : 5) | NC score | 0.989015 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16331 | Gene names | Pah | |||
|
Domain Architecture |
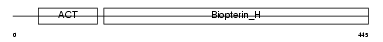
|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
PH4H_HUMAN
|
||||||
| θ value | 4.90052e-139 (rank : 6) | NC score | 0.988778 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00439, Q16717, Q8TC14 | Gene names | PAH | |||
|
Domain Architecture |
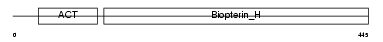
|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
TY3H_MOUSE
|
||||||
| θ value | 4.92331e-123 (rank : 7) | NC score | 0.983765 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24529 | Gene names | Th | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine 3-monooxygenase (EC 1.14.16.2) (Tyrosine 3-hydroxylase) (TH). | |||||
|
TY3H_HUMAN
|
||||||
| θ value | 1.02657e-120 (rank : 8) | NC score | 0.982214 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07101, Q15585, Q15588, Q15589 | Gene names | TH, TYH | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine 3-monooxygenase (EC 1.14.16.2) (Tyrosine 3-hydroxylase) (TH). | |||||
|
EMR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.019423 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.016914 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.012548 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.013055 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.012946 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.014399 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RENR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.018677 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75787, Q5QTQ7, Q6T7F5, Q8NBP3, Q8NG15, Q96FV6, Q96LB5, Q9H2P8, Q9UG89 | Gene names | ATP6AP2, ATP6IP2, CAPER, ELDF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Renin receptor precursor (Renin/prorenin receptor) (ATPase H(+)- transporting lysosomal accessory protein 2) (ATPase H(+)-transporting lysosomal-interacting protein 2) (Vacuolar ATP synthase membrane sector-associated protein M8-9) (V-ATPase M8.9 subunit) (ATP6M8-9) (N14F) (ER-localized type I transmembrane adaptor) (Embryonic liver differentiation factor 10). | |||||
|
Z297B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.001863 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 792 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43298 | Gene names | ZNF297B, KIAA0414, ZBTB22B | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 297B (ZnF-x) (Zinc finger and BTB domain- containing protein 22B). | |||||
|
STAG2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.009308 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
CBPB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.006320 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.004228 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
P55G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.003094 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
RENR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.016304 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CYN9, Q8BVU6, Q91YU5 | Gene names | Atp6ap2, Atp6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Renin receptor precursor (Renin/prorenin receptor) (ATPase H(+)- transporting lysosomal accessory protein 2) (ATPase H(+)-transporting lysosomal-interacting protein 2). | |||||
|
TPH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17752, O95188, O95189, Q16736 | Gene names | TPH1, TPH, TRPH | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
TPH1_MOUSE
|
||||||
| NC score | 0.999098 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17532 | Gene names | Tph1, Tph | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 1 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 1). | |||||
|
TPH2_HUMAN
|
||||||
| NC score | 0.996531 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWU9 | Gene names | TPH2, NTPH | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 2 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 2) (Neuronal tryptophan hydroxylase). | |||||
|
TPH2_MOUSE
|
||||||
| NC score | 0.996424 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGV2 | Gene names | Tph2, Ntph | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophan 5-hydroxylase 2 (EC 1.14.16.4) (Tryptophan 5-monooxygenase 2) (Neuronal tryptophan hydroxylase). | |||||
|
PH4H_MOUSE
|
||||||
| NC score | 0.989015 (rank : 5) | θ value | 5.78879e-140 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16331 | Gene names | Pah | |||
|
Domain Architecture |

|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
PH4H_HUMAN
|
||||||
| NC score | 0.988778 (rank : 6) | θ value | 4.90052e-139 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00439, Q16717, Q8TC14 | Gene names | PAH | |||
|
Domain Architecture |

|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
TY3H_MOUSE
|
||||||
| NC score | 0.983765 (rank : 7) | θ value | 4.92331e-123 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24529 | Gene names | Th | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine 3-monooxygenase (EC 1.14.16.2) (Tyrosine 3-hydroxylase) (TH). | |||||
|
TY3H_HUMAN
|
||||||
| NC score | 0.982214 (rank : 8) | θ value | 1.02657e-120 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07101, Q15585, Q15588, Q15589 | Gene names | TH, TYH | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine 3-monooxygenase (EC 1.14.16.2) (Tyrosine 3-hydroxylase) (TH). | |||||
|
EMR1_HUMAN
|
||||||
| NC score | 0.019423 (rank : 9) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
RENR_HUMAN
|
||||||
| NC score | 0.018677 (rank : 10) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75787, Q5QTQ7, Q6T7F5, Q8NBP3, Q8NG15, Q96FV6, Q96LB5, Q9H2P8, Q9UG89 | Gene names | ATP6AP2, ATP6IP2, CAPER, ELDF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Renin receptor precursor (Renin/prorenin receptor) (ATPase H(+)- transporting lysosomal accessory protein 2) (ATPase H(+)-transporting lysosomal-interacting protein 2) (Vacuolar ATP synthase membrane sector-associated protein M8-9) (V-ATPase M8.9 subunit) (ATP6M8-9) (N14F) (ER-localized type I transmembrane adaptor) (Embryonic liver differentiation factor 10). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.016914 (rank : 11) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
RENR_MOUSE
|
||||||
| NC score | 0.016304 (rank : 12) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CYN9, Q8BVU6, Q91YU5 | Gene names | Atp6ap2, Atp6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Renin receptor precursor (Renin/prorenin receptor) (ATPase H(+)- transporting lysosomal accessory protein 2) (ATPase H(+)-transporting lysosomal-interacting protein 2). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.014399 (rank : 13) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.013055 (rank : 14) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.012946 (rank : 15) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.012548 (rank : 16) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
STAG2_MOUSE
|
||||||
| NC score | 0.009308 (rank : 17) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35638, Q6NZN7 | Gene names | Stag2, Sa2, Sap2 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-2 (Stromal antigen 2) (SCC3 homolog 2). | |||||
|
CBPB2_HUMAN
|
||||||
| NC score | 0.006320 (rank : 18) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IY4, Q15114, Q5T9K1, Q5T9K2, Q9P2Y6 | Gene names | CPB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase B2 precursor (EC 3.4.17.20) (Carboxypeptidase U) (CPU) (Thrombin-activable fibrinolysis inhibitor) (TAFI) (Plasma carboxypeptidase B) (pCPB). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.004228 (rank : 19) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.003094 (rank : 20) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
Z297B_HUMAN
|
||||||
| NC score | 0.001863 (rank : 21) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 792 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43298 | Gene names | ZNF297B, KIAA0414, ZBTB22B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297B (ZnF-x) (Zinc finger and BTB domain- containing protein 22B). | |||||