Please be patient as the page loads
|
SULF1_MOUSE
|
||||||
| SwissProt Accessions | Q8K007, Q8BLJ0, Q8C1D3 | Gene names | Sulf1 | |||
|
Domain Architecture |
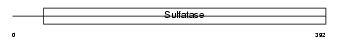
|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (MSulf-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SULF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995784 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IWU6, Q86YV8, Q8NCA2, Q9UPS5 | Gene names | SULF1, KIAA1077 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (HSulf-1). | |||||
|
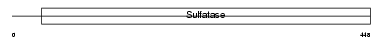
SULF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K007, Q8BLJ0, Q8C1D3 | Gene names | Sulf1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (MSulf-1). | |||||
|
SULF2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992538 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWU5, Q6UX86, Q96SG2, Q9H1H0, Q9UJR3, Q9ULH3 | Gene names | SULF2, KIAA1247 | |||
|
Domain Architecture |
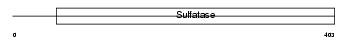
|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (HSulf-2). | |||||
|
SULF2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992310 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFG0, Q8BM68, Q8BUZ4, Q8BX28, Q8BZ51, Q8C169, Q9D8E2 | Gene names | Sulf2 | |||
|
Domain Architecture |
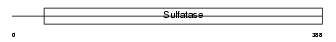
|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (MSulf-2). | |||||
|
GL6S_HUMAN
|
||||||
| θ value | 8.20983e-86 (rank : 5) | NC score | 0.940430 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15586 | Gene names | GNS | |||
|
Domain Architecture |
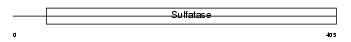
|
|||||
| Description | N-acetylglucosamine-6-sulfatase precursor (EC 3.1.6.14) (G6S) (Glucosamine-6-sulfatase). | |||||
|
ARSK_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 6) | NC score | 0.708321 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6UWY0, Q8N3Q8 | Gene names | ARSK, TSULF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase K precursor (EC 3.1.6.-) (ASK) (Telethon sulfatase). | |||||
|
ARSA_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 7) | NC score | 0.543593 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P50428 | Gene names | Arsa, As2 | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase). | |||||
|
IDS_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 8) | NC score | 0.572773 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22304, Q14604 | Gene names | IDS | |||
|
Domain Architecture |
|
|||||
| Description | Iduronate 2-sulfatase precursor (EC 3.1.6.13) (Alpha-L-iduronate sulfate sulfatase) (Idursulfase) [Contains: Iduronate 2-sulfatase 42 kDa chain; Iduronate 2-sulfatase 14 kDa chain]. | |||||
|
IDS_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 9) | NC score | 0.576594 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q08890, Q3TM30 | Gene names | Ids | |||
|
Domain Architecture |
|
|||||
| Description | Iduronate 2-sulfatase precursor (EC 3.1.6.13) (Alpha-L-iduronate sulfate sulfatase). | |||||
|
GA6S_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 10) | NC score | 0.550384 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P34059, Q86VK3 | Gene names | GALNS | |||
|
Domain Architecture |
|
|||||
| Description | N-acetylgalactosamine-6-sulfatase precursor (EC 3.1.6.4) (N- acetylgalactosamine-6-sulfate sulfatase) (Galactose-6-sulfate sulfatase) (GalNAc6S sulfatase) (Chondroitinsulfatase) (Chondroitinase). | |||||
|
ARSJ_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 11) | NC score | 0.503731 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FYB0, Q5FWE4, Q6UWT9 | Gene names | ARSJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSJ_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 12) | NC score | 0.500429 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM89, Q8BKG5, Q8BL46 | Gene names | Arsj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSI_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 13) | NC score | 0.498255 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FYB1 | Gene names | ARSI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase I precursor (EC 3.1.6.-) (ASI). | |||||
|
SPHM_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 14) | NC score | 0.570549 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51688 | Gene names | SGSH, HSS | |||
|
Domain Architecture |
|
|||||
| Description | N-sulphoglucosamine sulphohydrolase precursor (EC 3.10.1.1) (Sulfoglucosamine sulfamidase) (Sulphamidase). | |||||
|
ARSG_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 15) | NC score | 0.497857 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3TYD4, Q5XFU5, Q69ZT6, Q8CHS3, Q8VBZ5, Q9D3B4 | Gene names | Arsg, Kiaa1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
ARSB_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 16) | NC score | 0.485250 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15848 | Gene names | ARSB | |||
|
Domain Architecture |
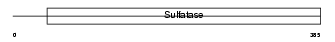
|
|||||
| Description | Arylsulfatase B precursor (EC 3.1.6.12) (ASB) (N-acetylgalactosamine- 4-sulfatase) (G4S). | |||||
|
STS_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 17) | NC score | 0.458657 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08842 | Gene names | STS | |||
|
Domain Architecture |
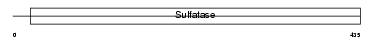
|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
STS_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 18) | NC score | 0.438328 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50427 | Gene names | Sts | |||
|
Domain Architecture |
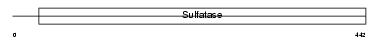
|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ARSF_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 19) | NC score | 0.430093 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54793, Q8TCC5 | Gene names | ARSF | |||
|
Domain Architecture |
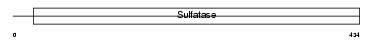
|
|||||
| Description | Arylsulfatase F precursor (EC 3.1.6.-) (ASF). | |||||
|
ARSA_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 20) | NC score | 0.495712 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15289, Q6ICI5, Q96CJ0 | Gene names | ARSA | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase) [Contains: Arylsulfatase A component B; Arylsulfatase A component C]. | |||||
|
ARSD_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 21) | NC score | 0.413782 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51689, Q9UHJ8 | Gene names | ARSD | |||
|
Domain Architecture |
|
|||||
| Description | Arylsulfatase D precursor (EC 3.1.6.-) (ASD). | |||||
|
ARSG_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 22) | NC score | 0.446981 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96EG1, Q6UXF2, Q9Y2K4 | Gene names | ARSG, KIAA1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
ARSE_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 23) | NC score | 0.408854 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51690, Q53FT2, Q53FU8 | Gene names | ARSE | |||
|
Domain Architecture |
|
|||||
| Description | Arylsulfatase E precursor (EC 3.1.6.-) (ASE). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.019105 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ZFP46_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.006202 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03309 | Gene names | Zfp46, Mlz-4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 46 (Zfp-46) (Zinc finger protein MLZ-4). | |||||
|
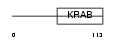
ZN436_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.006194 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C0F3, Q658I9 | Gene names | ZNF436, KIAA1710 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 436. | |||||
|
ZN436_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.006239 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||
|
ZN565_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.005453 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N9K5, Q6NUS2 | Gene names | ZNF565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 565. | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.039212 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
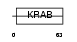
ZN347_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.005538 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96SE7, Q8TCN1 | Gene names | ZNF347, ZNF1111 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 347 (Zinc finger protein 1111). | |||||
|
CF170_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.027863 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.007652 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.020412 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
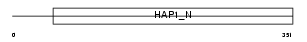
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.011078 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
TCF17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.004826 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60765, Q9UNJ8 | Gene names | ZNF354A, EZNF, HKL1, TCF17 | |||
|
Domain Architecture |
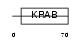
|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Zinc finger protein eZNF). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.007502 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.024065 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.020426 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.010855 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.007847 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
TNR11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.014645 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |
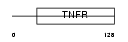
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
ZN479_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.004522 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JC4 | Gene names | ZNF479 | |||
|
Domain Architecture |
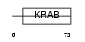
|
|||||
| Description | Zinc finger protein 479 (Zinc finger protein Kr19) (HKr19). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.011679 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ZFP30_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.004612 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60585, Q9CRV9 | Gene names | Zfp30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 (Zfp-30). | |||||
|
ZN655_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.004901 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N720, Q8IV00, Q8TA89, Q96EZ3 | Gene names | ZNF655, VIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 655 (Vav-interacting Krueppel-like protein). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.001709 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.006950 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.004435 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
RO52_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.000928 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19474, Q96RF8 | Gene names | TRIM21, RNF81, RO52, SSA1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21) (RING finger protein 81). | |||||
|
SLIT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.000088 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
TEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.012878 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
ZFP14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.004467 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCL3 | Gene names | ZFP14, KIAA1559 | |||
|
Domain Architecture |
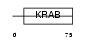
|
|||||
| Description | Zinc finger protein 14 homolog (Zfp-14). | |||||
|
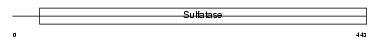
SULF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K007, Q8BLJ0, Q8C1D3 | Gene names | Sulf1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (MSulf-1). | |||||
|
SULF1_HUMAN
|
||||||
| NC score | 0.995784 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IWU6, Q86YV8, Q8NCA2, Q9UPS5 | Gene names | SULF1, KIAA1077 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (HSulf-1). | |||||
|
SULF2_HUMAN
|
||||||
| NC score | 0.992538 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWU5, Q6UX86, Q96SG2, Q9H1H0, Q9UJR3, Q9ULH3 | Gene names | SULF2, KIAA1247 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (HSulf-2). | |||||
|
SULF2_MOUSE
|
||||||
| NC score | 0.992310 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CFG0, Q8BM68, Q8BUZ4, Q8BX28, Q8BZ51, Q8C169, Q9D8E2 | Gene names | Sulf2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-2 precursor (EC 3.1.6.-) (MSulf-2). | |||||
|
GL6S_HUMAN
|
||||||
| NC score | 0.940430 (rank : 5) | θ value | 8.20983e-86 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15586 | Gene names | GNS | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylglucosamine-6-sulfatase precursor (EC 3.1.6.14) (G6S) (Glucosamine-6-sulfatase). | |||||
|
ARSK_HUMAN
|
||||||
| NC score | 0.708321 (rank : 6) | θ value | 1.63225e-17 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6UWY0, Q8N3Q8 | Gene names | ARSK, TSULF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase K precursor (EC 3.1.6.-) (ASK) (Telethon sulfatase). | |||||
|
IDS_MOUSE
|
||||||
| NC score | 0.576594 (rank : 7) | θ value | 1.2105e-12 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q08890, Q3TM30 | Gene names | Ids | |||
|
Domain Architecture |

|
|||||
| Description | Iduronate 2-sulfatase precursor (EC 3.1.6.13) (Alpha-L-iduronate sulfate sulfatase). | |||||
|
IDS_HUMAN
|
||||||
| NC score | 0.572773 (rank : 8) | θ value | 3.18553e-13 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22304, Q14604 | Gene names | IDS | |||
|
Domain Architecture |

|
|||||
| Description | Iduronate 2-sulfatase precursor (EC 3.1.6.13) (Alpha-L-iduronate sulfate sulfatase) (Idursulfase) [Contains: Iduronate 2-sulfatase 42 kDa chain; Iduronate 2-sulfatase 14 kDa chain]. | |||||
|
SPHM_HUMAN
|
||||||
| NC score | 0.570549 (rank : 9) | θ value | 6.21693e-09 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51688 | Gene names | SGSH, HSS | |||
|
Domain Architecture |

|
|||||
| Description | N-sulphoglucosamine sulphohydrolase precursor (EC 3.10.1.1) (Sulfoglucosamine sulfamidase) (Sulphamidase). | |||||
|
GA6S_HUMAN
|
||||||
| NC score | 0.550384 (rank : 10) | θ value | 1.58096e-12 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P34059, Q86VK3 | Gene names | GALNS | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylgalactosamine-6-sulfatase precursor (EC 3.1.6.4) (N- acetylgalactosamine-6-sulfate sulfatase) (Galactose-6-sulfate sulfatase) (GalNAc6S sulfatase) (Chondroitinsulfatase) (Chondroitinase). | |||||
|
ARSA_MOUSE
|
||||||
| NC score | 0.543593 (rank : 11) | θ value | 2.43908e-13 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P50428 | Gene names | Arsa, As2 | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase). | |||||
|
ARSJ_HUMAN
|
||||||
| NC score | 0.503731 (rank : 12) | θ value | 5.08577e-11 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FYB0, Q5FWE4, Q6UWT9 | Gene names | ARSJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSJ_MOUSE
|
||||||
| NC score | 0.500429 (rank : 13) | θ value | 1.9326e-10 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM89, Q8BKG5, Q8BL46 | Gene names | Arsj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSI_HUMAN
|
||||||
| NC score | 0.498255 (rank : 14) | θ value | 4.30538e-10 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5FYB1 | Gene names | ARSI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase I precursor (EC 3.1.6.-) (ASI). | |||||
|
ARSG_MOUSE
|
||||||
| NC score | 0.497857 (rank : 15) | θ value | 1.80886e-08 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3TYD4, Q5XFU5, Q69ZT6, Q8CHS3, Q8VBZ5, Q9D3B4 | Gene names | Arsg, Kiaa1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
ARSA_HUMAN
|
||||||
| NC score | 0.495712 (rank : 16) | θ value | 8.40245e-06 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15289, Q6ICI5, Q96CJ0 | Gene names | ARSA | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase) [Contains: Arylsulfatase A component B; Arylsulfatase A component C]. | |||||
|
ARSB_HUMAN
|
||||||
| NC score | 0.485250 (rank : 17) | θ value | 2.36244e-08 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15848 | Gene names | ARSB | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase B precursor (EC 3.1.6.12) (ASB) (N-acetylgalactosamine- 4-sulfatase) (G4S). | |||||
|
STS_HUMAN
|
||||||
| NC score | 0.458657 (rank : 18) | θ value | 1.99992e-07 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08842 | Gene names | STS | |||
|
Domain Architecture |

|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ARSG_HUMAN
|
||||||
| NC score | 0.446981 (rank : 19) | θ value | 5.44631e-05 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96EG1, Q6UXF2, Q9Y2K4 | Gene names | ARSG, KIAA1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
STS_MOUSE
|
||||||
| NC score | 0.438328 (rank : 20) | θ value | 4.45536e-07 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50427 | Gene names | Sts | |||
|
Domain Architecture |

|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ARSF_HUMAN
|
||||||
| NC score | 0.430093 (rank : 21) | θ value | 6.43352e-06 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54793, Q8TCC5 | Gene names | ARSF | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase F precursor (EC 3.1.6.-) (ASF). | |||||
|
ARSD_HUMAN
|
||||||
| NC score | 0.413782 (rank : 22) | θ value | 3.19293e-05 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51689, Q9UHJ8 | Gene names | ARSD | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase D precursor (EC 3.1.6.-) (ASD). | |||||
|
ARSE_HUMAN
|
||||||
| NC score | 0.408854 (rank : 23) | θ value | 7.1131e-05 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51690, Q53FT2, Q53FU8 | Gene names | ARSE | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase E precursor (EC 3.1.6.-) (ASE). | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.039212 (rank : 24) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
CF170_HUMAN
|
||||||
| NC score | 0.027863 (rank : 25) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.024065 (rank : 26) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.020426 (rank : 27) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.020412 (rank : 28) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.019105 (rank : 29) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
TNR11_HUMAN
|
||||||
| NC score | 0.014645 (rank : 30) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TEP1_HUMAN
|
||||||
| NC score | 0.012878 (rank : 31) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.011679 (rank : 32) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.011078 (rank : 33) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.010855 (rank : 34) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.007847 (rank : 35) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.007652 (rank : 36) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.007502 (rank : 37) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.006950 (rank : 38) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ZN436_MOUSE
|
||||||
| NC score | 0.006239 (rank : 39) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||
|
ZFP46_MOUSE
|
||||||
| NC score | 0.006202 (rank : 40) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03309 | Gene names | Zfp46, Mlz-4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 46 (Zfp-46) (Zinc finger protein MLZ-4). | |||||
|
ZN436_HUMAN
|
||||||
| NC score | 0.006194 (rank : 41) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C0F3, Q658I9 | Gene names | ZNF436, KIAA1710 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 436. | |||||
|
ZN347_HUMAN
|
||||||
| NC score | 0.005538 (rank : 42) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96SE7, Q8TCN1 | Gene names | ZNF347, ZNF1111 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 347 (Zinc finger protein 1111). | |||||
|
ZN565_HUMAN
|
||||||
| NC score | 0.005453 (rank : 43) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N9K5, Q6NUS2 | Gene names | ZNF565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 565. | |||||
|
ZN655_HUMAN
|
||||||
| NC score | 0.004901 (rank : 44) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N720, Q8IV00, Q8TA89, Q96EZ3 | Gene names | ZNF655, VIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 655 (Vav-interacting Krueppel-like protein). | |||||
|
TCF17_HUMAN
|
||||||
| NC score | 0.004826 (rank : 45) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60765, Q9UNJ8 | Gene names | ZNF354A, EZNF, HKL1, TCF17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Zinc finger protein eZNF). | |||||
|
ZFP30_MOUSE
|
||||||
| NC score | 0.004612 (rank : 46) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60585, Q9CRV9 | Gene names | Zfp30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 (Zfp-30). | |||||
|
ZN479_HUMAN
|
||||||
| NC score | 0.004522 (rank : 47) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JC4 | Gene names | ZNF479 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 479 (Zinc finger protein Kr19) (HKr19). | |||||
|
ZFP14_HUMAN
|
||||||
| NC score | 0.004467 (rank : 48) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCL3 | Gene names | ZFP14, KIAA1559 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 14 homolog (Zfp-14). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.004435 (rank : 49) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.001709 (rank : 50) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
RO52_HUMAN
|
||||||
| NC score | 0.000928 (rank : 51) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19474, Q96RF8 | Gene names | TRIM21, RNF81, RO52, SSA1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21) (RING finger protein 81). | |||||
|
SLIT3_HUMAN
|
||||||
| NC score | 0.000088 (rank : 52) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||