Please be patient as the page loads
|
S3TC1_HUMAN
|
||||||
| SwissProt Accessions | Q8TE82 | Gene names | SH3TC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S3TC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TE82 | Gene names | SH3TC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 1. | |||||
|
S3TC2_MOUSE
|
||||||
| θ value | 2.56852e-180 (rank : 2) | NC score | 0.924433 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80VA5, Q8BWH2, Q8BXB8, Q8C0G6 | Gene names | Sh3tc2, Kiaa1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
S3TC2_HUMAN
|
||||||
| θ value | 2.17438e-179 (rank : 3) | NC score | 0.928880 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TF17, Q9H8I5 | Gene names | SH3TC2, KIAA1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
GPSM1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 4) | NC score | 0.299154 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 5) | NC score | 0.303308 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 6) | NC score | 0.290639 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.287130 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.279635 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 9) | NC score | 0.271081 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.110115 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
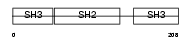
GRAP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.066767 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein. | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.102761 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.041212 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.025368 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.021469 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
GRAP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.061985 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
SRP68_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.052413 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHB9, Q8NCJ4, Q8WUK2 | Gene names | SRP68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal recognition particle 68 kDa protein (SRP68). | |||||
|
VAV3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.053730 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
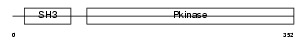
M3K10_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.016417 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
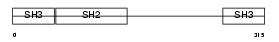
GRAP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.067106 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O89100 | Gene names | Grap2, Gads, Grb2l, Grid, Mona | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adaptor protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (GRB-2-like protein) (GRB2L) (Hematopoietic cell-associated adaptor protein GrpL) (GRB-2-related monocytic adapter protein) (Monocytic adapter) (MONA) (Adapter protein GRID). | |||||
|
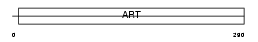
NAR3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.037143 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13508, Q8WVJ7, Q93069, Q96HL1 | Gene names | ART3, TMART | |||
|
Domain Architecture |
|
|||||
| Description | Ecto-ADP-ribosyltransferase 3 precursor (EC 2.4.2.31) (NAD(P)(+)-- arginine ADP-ribosyltransferase 3) (Mono(ADP-ribosyl)transferase 3). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.057799 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PEX5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.057459 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.015959 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
RAPSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.093255 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.045081 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
GRAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.058484 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75791, O43726 | Gene names | GRAP2, GADS, GRB2L, GRID | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (Grf40 adapter protein) (Grf-40) (GRB-2-like protein) (GRB2L) (GRBX) (P38) (Hematopoietic cell-associated adapter protein GrpL) (Adapter protein GRID) (SH3-SH2-SH3 adapter Mona). | |||||
|
M3KL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.019636 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCX8, Q5TCX7, Q5TCX9, Q8WWN1, Q8WWN2, Q96JM1 | Gene names | MLK4, KIAA1804 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase (EC 2.7.11.25) (Mixed lineage kinase 4). | |||||
|
PEX5R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.048971 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.049159 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.051239 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
DCTN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.037912 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJW0, Q8TAN8 | Gene names | DCTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
DCTN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.037915 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY8, Q923A0, Q9D4X0 | Gene names | Dctn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.055632 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.055752 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.026009 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
LRIG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.003683 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.029548 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
X3CL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.025245 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.011894 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
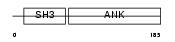
OSTF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.039824 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92882, Q5W126, Q96IJ4 | Gene names | OSTF1 | |||
|
Domain Architecture |
|
|||||
| Description | Osteoclast-stimulating factor 1. | |||||
|
OSTF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.034653 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62422, Q3UF05 | Gene names | Ostf1, Sh3d3, Sh3p2 | |||
|
Domain Architecture |
|
|||||
| Description | Osteoclast-stimulating factor 1 (SH3 domain protein 3). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.016070 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TTC7B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.020559 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
VAV2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.048742 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
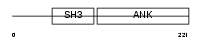
CBPA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.009332 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
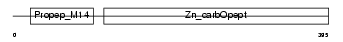
CRKL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.045936 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47941, Q3TQ18, Q8BGC5 | Gene names | Crkl, Crkol | |||
|
Domain Architecture |
|
|||||
| Description | Crk-like protein. | |||||
|
CRK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.049613 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46108 | Gene names | CRK | |||
|
Domain Architecture |
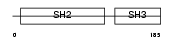
|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
CRK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.050333 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64010 | Gene names | Crk, Crko | |||
|
Domain Architecture |
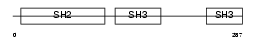
|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.023937 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.036872 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
PEX5R_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.045718 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
SNAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.030102 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H115, Q4G187, Q5JXF9, Q8N3C4 | Gene names | NAPB, SNAPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-soluble NSF attachment protein (SNAP-beta) (N-ethylmaleimide- sensitive factor attachment protein, beta). | |||||
|
SNAB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.030102 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28663 | Gene names | Napb, Snapb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-soluble NSF attachment protein (SNAP-beta) (N-ethylmaleimide- sensitive factor attachment protein, beta) (Brain protein I47). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.018350 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.013113 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TXNL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.016706 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |
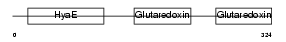
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
VAV3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.046799 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.010437 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CXB6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.005894 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70689 | Gene names | Gjb6, Cxn-30 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction beta-6 protein (Connexin-30) (Cx30). | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.009996 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
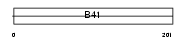
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
GUC2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.024952 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02747 | Gene names | GUCA2A, GUCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylin precursor (Guanylate cyclase activator 2A) (Guanylate cyclase-activating protein 1) (Gap-I) [Contains: HMW-guanylin; Guanylin]. | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.009145 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
MCF2L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.026133 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009112 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
PPIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.040792 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43586, O43585, O95657 | Gene names | PSTPIP1, CD2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 1 (PEST phosphatase-interacting protein 1) (CD2-binding protein 1) (H-PIP). | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.014046 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
VAV2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.047495 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.004353 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
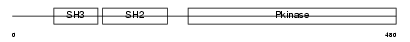
BLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.009268 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16277 | Gene names | Blk | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.043935 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
FA8A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.019404 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU6 | Gene names | FAM8A1, AHCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM8A1 (Autosomal highly conserved protein). | |||||
|
NFIP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.016579 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
PA24B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.008187 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
RGMA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.009459 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B86, Q9H0E6 | Gene names | RGMA, RGM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repulsive guidance molecule A precursor (RGM domain family member A). | |||||
|
SNAA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.027883 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54920, Q96IK3, Q9BVJ3 | Gene names | NAPA, SNAPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-soluble NSF attachment protein (SNAP-alpha) (N-ethylmaleimide- sensitive factor attachment protein, alpha). | |||||
|
SNAA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.027165 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DB05, Q543I3 | Gene names | Napa, Snapa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-soluble NSF attachment protein (SNAP-alpha) (N-ethylmaleimide- sensitive factor attachment protein, alpha). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.034024 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
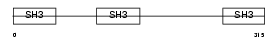
CD2AP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053137 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
CDC27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050660 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
GPSM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.069654 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4H4 | Gene names | GPSM3, AGS4, C6orf9, G18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 3 (Activator of G-protein signaling 4) (Protein G18) (G18.1b). | |||||
|
GPSM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.058716 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U1Z5, Q3U1M8, Q3UJZ5, Q8VDF9 | Gene names | Gpsm3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 3. | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051696 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
PCP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.080525 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVA1, Q3KRG7 | Gene names | PCP2 | |||
|
Domain Architecture |
|
|||||
| Description | Purkinje cell protein 2 homolog. | |||||
|
PCP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.076697 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12660, P22941, Q3TV53 | Gene names | Pcp2, Pcp-2 | |||
|
Domain Architecture |
|
|||||
| Description | Purkinje cell protein 2 (Protein PCD-5) (Purkinje cell-specific protein L7). | |||||
|
RAPSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.076661 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
SH3K1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050428 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050894 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
S3TC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TE82 | Gene names | SH3TC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 1. | |||||
|
S3TC2_HUMAN
|
||||||
| NC score | 0.928880 (rank : 2) | θ value | 2.17438e-179 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TF17, Q9H8I5 | Gene names | SH3TC2, KIAA1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
S3TC2_MOUSE
|
||||||
| NC score | 0.924433 (rank : 3) | θ value | 2.56852e-180 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80VA5, Q8BWH2, Q8BXB8, Q8C0G6 | Gene names | Sh3tc2, Kiaa1985 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain and tetratricopeptide repeats-containing protein 2. | |||||
|
GPSM1_MOUSE
|
||||||
| NC score | 0.303308 (rank : 4) | θ value | 1.43324e-05 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_HUMAN
|
||||||
| NC score | 0.299154 (rank : 5) | θ value | 2.88788e-06 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM2_HUMAN
|
||||||
| NC score | 0.290639 (rank : 6) | θ value | 5.44631e-05 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.287130 (rank : 7) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.279635 (rank : 8) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.271081 (rank : 9) | θ value | 0.000602161 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.110115 (rank : 10) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.102761 (rank : 11) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
RAPSN_MOUSE
|
||||||
| NC score | 0.093255 (rank : 12) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
PCP2_HUMAN
|
||||||
| NC score | 0.080525 (rank : 13) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVA1, Q3KRG7 | Gene names | PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Purkinje cell protein 2 homolog. | |||||
|
PCP2_MOUSE
|
||||||
| NC score | 0.076697 (rank : 14) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12660, P22941, Q3TV53 | Gene names | Pcp2, Pcp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Purkinje cell protein 2 (Protein PCD-5) (Purkinje cell-specific protein L7). | |||||
|
RAPSN_HUMAN
|
||||||
| NC score | 0.076661 (rank : 15) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
GPSM3_HUMAN
|
||||||
| NC score | 0.069654 (rank : 16) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4H4 | Gene names | GPSM3, AGS4, C6orf9, G18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 3 (Activator of G-protein signaling 4) (Protein G18) (G18.1b). | |||||
|
GRAP2_MOUSE
|
||||||
| NC score | 0.067106 (rank : 17) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O89100 | Gene names | Grap2, Gads, Grb2l, Grid, Mona | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adaptor protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (GRB-2-like protein) (GRB2L) (Hematopoietic cell-associated adaptor protein GrpL) (GRB-2-related monocytic adapter protein) (Monocytic adapter) (MONA) (Adapter protein GRID). | |||||
|
GRAP_HUMAN
|
||||||
| NC score | 0.066767 (rank : 18) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRAP_MOUSE
|
||||||
| NC score | 0.061985 (rank : 19) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
GPSM3_MOUSE
|
||||||
| NC score | 0.058716 (rank : 20) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U1Z5, Q3U1M8, Q3UJZ5, Q8VDF9 | Gene names | Gpsm3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 3. | |||||
|
GRAP2_HUMAN
|
||||||
| NC score | 0.058484 (rank : 21) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75791, O43726 | Gene names | GRAP2, GADS, GRB2L, GRID | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (Grf40 adapter protein) (Grf-40) (GRB-2-like protein) (GRB2L) (GRBX) (P38) (Hematopoietic cell-associated adapter protein GrpL) (Adapter protein GRID) (SH3-SH2-SH3 adapter Mona). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.057799 (rank : 22) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
PEX5_MOUSE
|
||||||
| NC score | 0.057459 (rank : 23) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09012 | Gene names | Pex5, Pxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor) (PXR1P) (PTS1R). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.055752 (rank : 24) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.055632 (rank : 25) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.053730 (rank : 26) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.053137 (rank : 27) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
SRP68_HUMAN
|
||||||
| NC score | 0.052413 (rank : 28) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHB9, Q8NCJ4, Q8WUK2 | Gene names | SRP68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal recognition particle 68 kDa protein (SRP68). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.051696 (rank : 29) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.051239 (rank : 30) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.050894 (rank : 31) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.050660 (rank : 32) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
SH3K1_HUMAN
|
||||||
| NC score | 0.050428 (rank : 33) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
CRK_MOUSE
|
||||||
| NC score | 0.050333 (rank : 34) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64010 | Gene names | Crk, Crko | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
CRK_HUMAN
|
||||||
| NC score | 0.049613 (rank : 35) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46108 | Gene names | CRK | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.049159 (rank : 36) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
PEX5R_HUMAN
|
||||||
| NC score | 0.048971 (rank : 37) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IYB4, Q9NQD1, Q9P2U3, Q9P2U4 | Gene names | PEX5L, PEX5R, PXR2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.048742 (rank : 38) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.047495 (rank : 39) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.046799 (rank : 40) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
CRKL_MOUSE
|
||||||
| NC score | 0.045936 (rank : 41) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47941, Q3TQ18, Q8BGC5 | Gene names | Crkl, Crkol | |||
|
Domain Architecture |

|
|||||
| Description | Crk-like protein. | |||||
|
PEX5R_MOUSE
|
||||||
| NC score | 0.045718 (rank : 42) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C437, Q8CA31, Q9DAA1, Q9JMB9 | Gene names | Pex5l, Pex2, Pex5r, Pxr2 | |||
|
Domain Architecture |

|
|||||
| Description | PEX5-related protein (Peroxin-5-related protein) (Pex5Rp) (PEX5-like protein) (PEX2-related protein). | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.045081 (rank : 43) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.043935 (rank : 44) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
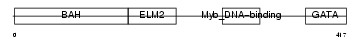
HRX_HUMAN
|
||||||
| NC score | 0.041212 (rank : 45) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PPIP1_HUMAN
|
||||||
| NC score | 0.040792 (rank : 46) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43586, O43585, O95657 | Gene names | PSTPIP1, CD2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-serine-threonine phosphatase-interacting protein 1 (PEST phosphatase-interacting protein 1) (CD2-binding protein 1) (H-PIP). | |||||
|
OSTF1_HUMAN
|
||||||
| NC score | 0.039824 (rank : 47) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92882, Q5W126, Q96IJ4 | Gene names | OSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteoclast-stimulating factor 1. | |||||
|
DCTN4_MOUSE
|
||||||
| NC score | 0.037915 (rank : 48) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY8, Q923A0, Q9D4X0 | Gene names | Dctn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
DCTN4_HUMAN
|
||||||
| NC score | 0.037912 (rank : 49) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJW0, Q8TAN8 | Gene names | DCTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
NAR3_HUMAN
|
||||||
| NC score | 0.037143 (rank : 50) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13508, Q8WVJ7, Q93069, Q96HL1 | Gene names | ART3, TMART | |||
|
Domain Architecture |

|
|||||
| Description | Ecto-ADP-ribosyltransferase 3 precursor (EC 2.4.2.31) (NAD(P)(+)-- arginine ADP-ribosyltransferase 3) (Mono(ADP-ribosyl)transferase 3). | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.036872 (rank : 51) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
OSTF1_MOUSE
|
||||||
| NC score | 0.034653 (rank : 52) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62422, Q3UF05 | Gene names | Ostf1, Sh3d3, Sh3p2 | |||
|
Domain Architecture |

|
|||||
| Description | Osteoclast-stimulating factor 1 (SH3 domain protein 3). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.034024 (rank : 53) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
SNAB_HUMAN
|
||||||
| NC score | 0.030102 (rank : 54) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H115, Q4G187, Q5JXF9, Q8N3C4 | Gene names | NAPB, SNAPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-soluble NSF attachment protein (SNAP-beta) (N-ethylmaleimide- sensitive factor attachment protein, beta). | |||||
|
SNAB_MOUSE
|
||||||
| NC score | 0.030102 (rank : 55) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28663 | Gene names | Napb, Snapb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-soluble NSF attachment protein (SNAP-beta) (N-ethylmaleimide- sensitive factor attachment protein, beta) (Brain protein I47). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.029548 (rank : 56) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
SNAA_HUMAN
|
||||||
| NC score | 0.027883 (rank : 57) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54920, Q96IK3, Q9BVJ3 | Gene names | NAPA, SNAPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-soluble NSF attachment protein (SNAP-alpha) (N-ethylmaleimide- sensitive factor attachment protein, alpha). | |||||
|
SNAA_MOUSE
|
||||||
| NC score | 0.027165 (rank : 58) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DB05, Q543I3 | Gene names | Napa, Snapa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-soluble NSF attachment protein (SNAP-alpha) (N-ethylmaleimide- sensitive factor attachment protein, alpha). | |||||
|
MCF2L_MOUSE
|
||||||
| NC score | 0.026133 (rank : 59) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.026009 (rank : 60) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.025368 (rank : 61) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
X3CL1_HUMAN
|
||||||
| NC score | 0.025245 (rank : 62) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
GUC2A_HUMAN
|
||||||
| NC score | 0.024952 (rank : 63) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02747 | Gene names | GUCA2A, GUCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylin precursor (Guanylate cyclase activator 2A) (Guanylate cyclase-activating protein 1) (Gap-I) [Contains: HMW-guanylin; Guanylin]. | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.023937 (rank : 64) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
M3K11_HUMAN
|
||||||
| NC score | 0.021469 (rank : 65) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
TTC7B_HUMAN
|
||||||
| NC score | 0.020559 (rank : 66) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
M3KL4_HUMAN
|
||||||
| NC score | 0.019636 (rank : 67) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5TCX8, Q5TCX7, Q5TCX9, Q8WWN1, Q8WWN2, Q96JM1 | Gene names | MLK4, KIAA1804 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase (EC 2.7.11.25) (Mixed lineage kinase 4). | |||||
|
FA8A1_HUMAN
|
||||||
| NC score | 0.019404 (rank : 68) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU6 | Gene names | FAM8A1, AHCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM8A1 (Autosomal highly conserved protein). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.018350 (rank : 69) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
TXNL2_HUMAN
|
||||||
| NC score | 0.016706 (rank : 70) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
NFIP2_HUMAN
|
||||||
| NC score | 0.016579 (rank : 71) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | 0.016417 (rank : 72) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.016070 (rank : 73) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.015959 (rank : 74) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.014046 (rank : 75) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.013113 (rank : 76) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.011894 (rank : 77) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.010437 (rank : 78) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.009996 (rank : 79) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
RGMA_HUMAN
|
||||||
| NC score | 0.009459 (rank : 80) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B86, Q9H0E6 | Gene names | RGMA, RGM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repulsive guidance molecule A precursor (RGM domain family member A). | |||||
|
CBPA3_HUMAN
|
||||||
| NC score | 0.009332 (rank : 81) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15088, Q96E94 | Gene names | CPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell carboxypeptidase A precursor (EC 3.4.17.1) (MC-CPA) (Carboxypeptidase A3). | |||||
|
BLK_MOUSE
|
||||||
| NC score | 0.009268 (rank : 82) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16277 | Gene names | Blk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.009145 (rank : 83) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.009112 (rank : 84) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
PA24B_MOUSE
|
||||||
| NC score | 0.008187 (rank : 85) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
CXB6_MOUSE
|
||||||
| NC score | 0.005894 (rank : 86) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70689 | Gene names | Gjb6, Cxn-30 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction beta-6 protein (Connexin-30) (Cx30). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.004353 (rank : 87) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
LRIG1_HUMAN
|
||||||
| NC score | 0.003683 (rank : 88) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||