Please be patient as the page loads
|
PPGB_MOUSE
|
||||||
| SwissProt Accessions | P16675, Q8VEF6 | Gene names | Ppgb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PPGB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997998 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
PPGB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16675, Q8VEF6 | Gene names | Ppgb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
CPVL_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 3) | NC score | 0.850307 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3G5, Q6UX20, Q8NBL7, Q96AR7, Q9HB41 | Gene names | CPVL, VLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine carboxypeptidase CPVL precursor (EC 3.4.16.-) (Carboxypeptidase, vitellogenic-like) (Vitellogenic carboxypeptidase- like protein) (VCP-like protein) (HVLP). | |||||
|
RISC_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 4) | NC score | 0.676128 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920A5 | Gene names | Scpep1, Risc | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
RISC_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 5) | NC score | 0.637225 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HB40, Q96A94, Q9H3F0 | Gene names | SCPEP1, RISC, SCP1 | |||
|
Domain Architecture |
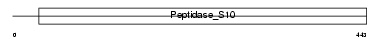
|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
POMT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.035198 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKY4, Q9NSG6, Q9P1W0, Q9P1W2 | Gene names | POMT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
POMT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.033889 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ4, Q8R4Z0 | Gene names | Pomt2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
KLF5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.002636 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.010125 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
PPGB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16675, Q8VEF6 | Gene names | Ppgb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
PPGB_HUMAN
|
||||||
| NC score | 0.997998 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
CPVL_HUMAN
|
||||||
| NC score | 0.850307 (rank : 3) | θ value | 1.08474e-45 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3G5, Q6UX20, Q8NBL7, Q96AR7, Q9HB41 | Gene names | CPVL, VLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine carboxypeptidase CPVL precursor (EC 3.4.16.-) (Carboxypeptidase, vitellogenic-like) (Vitellogenic carboxypeptidase- like protein) (VCP-like protein) (HVLP). | |||||
|
RISC_MOUSE
|
||||||
| NC score | 0.676128 (rank : 4) | θ value | 5.42112e-21 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920A5 | Gene names | Scpep1, Risc | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
RISC_HUMAN
|
||||||
| NC score | 0.637225 (rank : 5) | θ value | 3.40345e-15 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HB40, Q96A94, Q9H3F0 | Gene names | SCPEP1, RISC, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
POMT2_HUMAN
|
||||||
| NC score | 0.035198 (rank : 6) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKY4, Q9NSG6, Q9P1W0, Q9P1W2 | Gene names | POMT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
POMT2_MOUSE
|
||||||
| NC score | 0.033889 (rank : 7) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ4, Q8R4Z0 | Gene names | Pomt2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.010125 (rank : 8) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
KLF5_MOUSE
|
||||||
| NC score | 0.002636 (rank : 9) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||