Please be patient as the page loads
|
RISC_MOUSE
|
||||||
| SwissProt Accessions | Q920A5 | Gene names | Scpep1, Risc | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RISC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996083 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HB40, Q96A94, Q9H3F0 | Gene names | SCPEP1, RISC, SCP1 | |||
|
Domain Architecture |
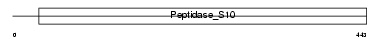
|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
RISC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920A5 | Gene names | Scpep1, Risc | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
CPVL_HUMAN
|
||||||
| θ value | 2.05049e-36 (rank : 3) | NC score | 0.817003 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3G5, Q6UX20, Q8NBL7, Q96AR7, Q9HB41 | Gene names | CPVL, VLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine carboxypeptidase CPVL precursor (EC 3.4.16.-) (Carboxypeptidase, vitellogenic-like) (Vitellogenic carboxypeptidase- like protein) (VCP-like protein) (HVLP). | |||||
|
PPGB_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 4) | NC score | 0.686454 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
PPGB_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 5) | NC score | 0.676128 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16675, Q8VEF6 | Gene names | Ppgb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.022173 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
KPTN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.038735 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCX6 | Gene names | Kptn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kaptin. | |||||
|
AT131_MOUSE
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.027814 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
IF2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.044521 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05198 | Gene names | EIF2S1, EIF2A | |||
|
Domain Architecture |
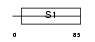
|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 1 (Eukaryotic translation initiation factor 2 subunit alpha) (eIF-2-alpha) (EIF- 2alpha) (EIF-2A). | |||||
|
IF2A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.044631 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWX6, Q3TIQ0 | Gene names | Eif2s1, Eif2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 1 (Eukaryotic translation initiation factor 2 subunit alpha) (eIF-2-alpha) (EIF- 2alpha) (EIF-2A). | |||||
|
AT131_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.024312 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.005696 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
RISC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920A5 | Gene names | Scpep1, Risc | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
RISC_HUMAN
|
||||||
| NC score | 0.996083 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HB40, Q96A94, Q9H3F0 | Gene names | SCPEP1, RISC, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoid-inducible serine carboxypeptidase precursor (EC 3.4.16.-) (Serine carboxypeptidase 1). | |||||
|
CPVL_HUMAN
|
||||||
| NC score | 0.817003 (rank : 3) | θ value | 2.05049e-36 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3G5, Q6UX20, Q8NBL7, Q96AR7, Q9HB41 | Gene names | CPVL, VLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine carboxypeptidase CPVL precursor (EC 3.4.16.-) (Carboxypeptidase, vitellogenic-like) (Vitellogenic carboxypeptidase- like protein) (VCP-like protein) (HVLP). | |||||
|
PPGB_HUMAN
|
||||||
| NC score | 0.686454 (rank : 4) | θ value | 1.6852e-22 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
PPGB_MOUSE
|
||||||
| NC score | 0.676128 (rank : 5) | θ value | 5.42112e-21 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16675, Q8VEF6 | Gene names | Ppgb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
IF2A_MOUSE
|
||||||
| NC score | 0.044631 (rank : 6) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWX6, Q3TIQ0 | Gene names | Eif2s1, Eif2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 1 (Eukaryotic translation initiation factor 2 subunit alpha) (eIF-2-alpha) (EIF- 2alpha) (EIF-2A). | |||||
|
IF2A_HUMAN
|
||||||
| NC score | 0.044521 (rank : 7) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05198 | Gene names | EIF2S1, EIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 1 (Eukaryotic translation initiation factor 2 subunit alpha) (eIF-2-alpha) (EIF- 2alpha) (EIF-2A). | |||||
|
KPTN_MOUSE
|
||||||
| NC score | 0.038735 (rank : 8) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCX6 | Gene names | Kptn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kaptin. | |||||
|
AT131_MOUSE
|
||||||
| NC score | 0.027814 (rank : 9) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT131_HUMAN
|
||||||
| NC score | 0.024312 (rank : 10) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.022173 (rank : 11) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.005696 (rank : 12) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||