Please be patient as the page loads
|
OST1_MOUSE
|
||||||
| SwissProt Accessions | O35310 | Gene names | Hs3st1, 3ost, 3ost1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OST1_MOUSE
|
||||||
| θ value | 2.93876e-176 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35310 | Gene names | Hs3st1, 3ost, 3ost1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1). | |||||
|
OST1_HUMAN
|
||||||
| θ value | 1.12453e-151 (rank : 2) | NC score | 0.999289 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14792, Q6PEY8 | Gene names | HS3ST1, 3OST, 3OST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1) (h3-OST-1). | |||||
|
OST5_MOUSE
|
||||||
| θ value | 1.14825e-79 (rank : 3) | NC score | 0.986961 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BSL4 | Gene names | Hs3st5, Hs3ost5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5). | |||||
|
OST5_HUMAN
|
||||||
| θ value | 1.95861e-79 (rank : 4) | NC score | 0.986863 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZT8, Q52LI2, Q8N285 | Gene names | HS3ST5, 3OST5, HS3OST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5) (h3-OST-5). | |||||
|
OST4_HUMAN
|
||||||
| θ value | 1.19377e-60 (rank : 5) | NC score | 0.964131 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
OST2_HUMAN
|
||||||
| θ value | 2.65945e-60 (rank : 6) | NC score | 0.971097 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y278 | Gene names | HS3ST2, 3OST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2) (h3-OST-2). | |||||
|
OST3A_MOUSE
|
||||||
| θ value | 2.65945e-60 (rank : 7) | NC score | 0.967300 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BKN6 | Gene names | Hs3st3a1, 3ost3a1, Hs3st3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1). | |||||
|
OST3B_MOUSE
|
||||||
| θ value | 2.65945e-60 (rank : 8) | NC score | 0.966274 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
OST2_MOUSE
|
||||||
| θ value | 3.47335e-60 (rank : 9) | NC score | 0.970222 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q673U1, Q8BLP1, Q8C055 | Gene names | Hs3st2, 3ost2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2). | |||||
|
OST6_MOUSE
|
||||||
| θ value | 2.25136e-59 (rank : 10) | NC score | 0.968948 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5GFD5 | Gene names | Hs3st6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (3-OST-6). | |||||
|
OST6_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 11) | NC score | 0.969000 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
OST3B_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 12) | NC score | 0.966870 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
OST3A_HUMAN
|
||||||
| θ value | 1.23536e-57 (rank : 13) | NC score | 0.968734 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y663 | Gene names | HS3ST3A1, 3OST3A1, HS3ST3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1) (h3-OST-3A). | |||||
|
NDST4_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 14) | NC score | 0.787813 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST3_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 15) | NC score | 0.789127 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95803, Q4W5C1, Q4W5D0, Q6UWC5, Q9UP21 | Gene names | NDST3, HSST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (hNDST-3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST4_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 16) | NC score | 0.785482 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQW8 | Gene names | Ndst4, Hsst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST2_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 17) | NC score | 0.786777 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST3_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 18) | NC score | 0.784865 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQH7, Q6AXE0, Q9D557 | Gene names | Ndst3, Hsst3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST2_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 19) | NC score | 0.783435 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52850, Q3UDF4, Q549P5 | Gene names | Ndst2, Hsst2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) (Mndns) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST1_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 20) | NC score | 0.772361 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHN9, O70353, Q3TBX3, Q3TDS3, Q8BZE5, Q9R206 | Gene names | Ndst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
NDST1_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 21) | NC score | 0.771930 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52848, Q96E57 | Gene names | NDST1, HSST, HSST1 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
ST4S6_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 22) | NC score | 0.437968 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LFX5, O60338, O60474, Q86VM4 | Gene names | GALNAC4S6ST, BRAG, KIAA0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein) (hBRAG). | |||||
|
ST4S6_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 23) | NC score | 0.455749 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XQ5, Q80TW4, Q8BLQ5, Q9D2N6 | Gene names | Galnac4s6st, Brag, Kiaa0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein). | |||||
|
GLPK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.041242 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14410 | Gene names | GK2, GKP2, GKTA | |||
|
Domain Architecture |
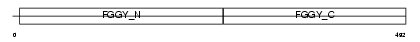
|
|||||
| Description | Glycerol kinase, testis specific 2 (EC 2.7.1.30) (ATP:glycerol 3- phosphotransferase) (Glycerokinase) (GK). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.003405 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
GLPK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.031950 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P32189, Q8IVR5, Q9UMP0, Q9UMP1 | Gene names | GK | |||
|
Domain Architecture |
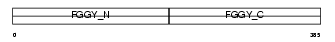
|
|||||
| Description | Glycerol kinase (EC 2.7.1.30) (ATP:glycerol 3-phosphotransferase) (Glycerokinase) (GK). | |||||
|
GLPK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.031948 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64516 | Gene names | Gk, Gyk | |||
|
Domain Architecture |
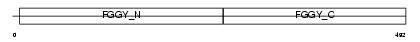
|
|||||
| Description | Glycerol kinase (EC 2.7.1.30) (ATP:glycerol 3-phosphotransferase) (Glycerokinase) (GK). | |||||
|
GLPK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.031385 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14409 | Gene names | GKP3, GKTB | |||
|
Domain Architecture |
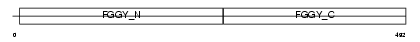
|
|||||
| Description | Glycerol kinase, testis specific 1 (EC 2.7.1.30) (ATP:glycerol 3- phosphotransferase) (Glycerokinase) (GK). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.005983 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
PYGM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.004050 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11217 | Gene names | PYGM | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
OST1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.93876e-176 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35310 | Gene names | Hs3st1, 3ost, 3ost1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1). | |||||
|
OST1_HUMAN
|
||||||
| NC score | 0.999289 (rank : 2) | θ value | 1.12453e-151 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14792, Q6PEY8 | Gene names | HS3ST1, 3OST, 3OST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 1 precursor (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 1) (Heparan sulfate 3-O-sulfotransferase 1) (h3-OST-1). | |||||
|
OST5_MOUSE
|
||||||
| NC score | 0.986961 (rank : 3) | θ value | 1.14825e-79 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BSL4 | Gene names | Hs3st5, Hs3ost5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5). | |||||
|
OST5_HUMAN
|
||||||
| NC score | 0.986863 (rank : 4) | θ value | 1.95861e-79 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZT8, Q52LI2, Q8N285 | Gene names | HS3ST5, 3OST5, HS3OST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 5 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 5) (Heparan sulfate 3-O-sulfotransferase 5) (h3-OST-5). | |||||
|
OST2_HUMAN
|
||||||
| NC score | 0.971097 (rank : 5) | θ value | 2.65945e-60 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y278 | Gene names | HS3ST2, 3OST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2) (h3-OST-2). | |||||
|
OST2_MOUSE
|
||||||
| NC score | 0.970222 (rank : 6) | θ value | 3.47335e-60 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q673U1, Q8BLP1, Q8C055 | Gene names | Hs3st2, 3ost2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 2 (EC 2.8.2.29) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 2) (Heparan sulfate 3-O-sulfotransferase 2). | |||||
|
OST6_HUMAN
|
||||||
| NC score | 0.969000 (rank : 7) | θ value | 8.55514e-59 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
OST6_MOUSE
|
||||||
| NC score | 0.968948 (rank : 8) | θ value | 2.25136e-59 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5GFD5 | Gene names | Hs3st6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (3-OST-6). | |||||
|
OST3A_HUMAN
|
||||||
| NC score | 0.968734 (rank : 9) | θ value | 1.23536e-57 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y663 | Gene names | HS3ST3A1, 3OST3A1, HS3ST3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1) (h3-OST-3A). | |||||
|
OST3A_MOUSE
|
||||||
| NC score | 0.967300 (rank : 10) | θ value | 2.65945e-60 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BKN6 | Gene names | Hs3st3a1, 3ost3a1, Hs3st3a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3A1 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1) (Heparan sulfate 3-O-sulfotransferase 3A1). | |||||
|
OST3B_HUMAN
|
||||||
| NC score | 0.966870 (rank : 11) | θ value | 4.24587e-58 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y662 | Gene names | HS3ST3B1, 3OST3B1, HS3ST3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (h3-OST-3B). | |||||
|
OST3B_MOUSE
|
||||||
| NC score | 0.966274 (rank : 12) | θ value | 2.65945e-60 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QZS6 | Gene names | Hs3st3b1, 3ost3b1, Hs3st3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 3B1 (EC 2.8.2.30) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3B1) (Heparan sulfate 3-O-sulfotransferase 3B1) (m3-OST-3B). | |||||
|
OST4_HUMAN
|
||||||
| NC score | 0.964131 (rank : 13) | θ value | 1.19377e-60 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y661, Q5QI42, Q8NDC2 | Gene names | HS3ST4, 3OST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 4 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4) (Heparan sulfate 3-O-sulfotransferase 4) (h3-OST-4). | |||||
|
NDST3_HUMAN
|
||||||
| NC score | 0.789127 (rank : 14) | θ value | 1.2049e-28 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95803, Q4W5C1, Q4W5D0, Q6UWC5, Q9UP21 | Gene names | NDST3, HSST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (hNDST-3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST4_HUMAN
|
||||||
| NC score | 0.787813 (rank : 15) | θ value | 1.85889e-29 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST2_HUMAN
|
||||||
| NC score | 0.786777 (rank : 16) | θ value | 5.97985e-28 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST4_MOUSE
|
||||||
| NC score | 0.785482 (rank : 17) | θ value | 2.05525e-28 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQW8 | Gene names | Ndst4, Hsst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
NDST3_MOUSE
|
||||||
| NC score | 0.784865 (rank : 18) | θ value | 5.06226e-27 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQH7, Q6AXE0, Q9D557 | Gene names | Ndst3, Hsst3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 3 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 3) (NDST- 3) (N-heparan sulfate sulfotransferase 3) (N-HSST 3) [Includes: Heparan sulfate N-deacetylase 3 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 3 (EC 2.8.2.-)]. | |||||
|
NDST2_MOUSE
|
||||||
| NC score | 0.783435 (rank : 19) | θ value | 1.12775e-26 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52850, Q3UDF4, Q549P5 | Gene names | Ndst2, Hsst2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) (Mndns) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N-sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
NDST1_MOUSE
|
||||||
| NC score | 0.772361 (rank : 20) | θ value | 1.5242e-23 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UHN9, O70353, Q3TBX3, Q3TDS3, Q8BZE5, Q9R206 | Gene names | Ndst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
NDST1_HUMAN
|
||||||
| NC score | 0.771930 (rank : 21) | θ value | 4.43474e-23 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52848, Q96E57 | Gene names | NDST1, HSST, HSST1 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 1) (NDST- 1) ([Heparan sulfate]-glucosamine N-sulfotransferase 1) (HSNST 1) (N- heparan sulfate sulfotransferase 1) (N-HSST 1) [Includes: Heparan sulfate N-deacetylase 1 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 1 (EC 2.8.2.-)]. | |||||
|
ST4S6_MOUSE
|
||||||
| NC score | 0.455749 (rank : 22) | θ value | 0.00175202 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XQ5, Q80TW4, Q8BLQ5, Q9D2N6 | Gene names | Galnac4s6st, Brag, Kiaa0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein). | |||||
|
ST4S6_HUMAN
|
||||||
| NC score | 0.437968 (rank : 23) | θ value | 0.00175202 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7LFX5, O60338, O60474, Q86VM4 | Gene names | GALNAC4S6ST, BRAG, KIAA0598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase (EC 2.8.2.33) (GalNAc4S-6ST) (B-cell RAG-associated gene protein) (hBRAG). | |||||
|
GLPK2_HUMAN
|
||||||
| NC score | 0.041242 (rank : 24) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14410 | Gene names | GK2, GKP2, GKTA | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol kinase, testis specific 2 (EC 2.7.1.30) (ATP:glycerol 3- phosphotransferase) (Glycerokinase) (GK). | |||||
|
GLPK_HUMAN
|
||||||
| NC score | 0.031950 (rank : 25) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P32189, Q8IVR5, Q9UMP0, Q9UMP1 | Gene names | GK | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol kinase (EC 2.7.1.30) (ATP:glycerol 3-phosphotransferase) (Glycerokinase) (GK). | |||||
|
GLPK_MOUSE
|
||||||
| NC score | 0.031948 (rank : 26) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64516 | Gene names | Gk, Gyk | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol kinase (EC 2.7.1.30) (ATP:glycerol 3-phosphotransferase) (Glycerokinase) (GK). | |||||
|
GLPK3_HUMAN
|
||||||
| NC score | 0.031385 (rank : 27) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14409 | Gene names | GKP3, GKTB | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol kinase, testis specific 1 (EC 2.7.1.30) (ATP:glycerol 3- phosphotransferase) (Glycerokinase) (GK). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.005983 (rank : 28) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
PYGM_HUMAN
|
||||||
| NC score | 0.004050 (rank : 29) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11217 | Gene names | PYGM | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.003405 (rank : 30) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||