Please be patient as the page loads
|
NUD15_MOUSE
|
||||||
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUD15_MOUSE
|
||||||
| θ value | 6.93392e-93 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD15_HUMAN
|
||||||
| θ value | 9.7205e-79 (rank : 2) | NC score | 0.990451 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
8ODP_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.431314 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |
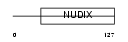
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 4) | NC score | 0.406858 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |
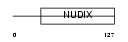
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
NUD13_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 5) | NC score | 0.229294 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD17_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 6) | NC score | 0.225216 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CWD3, Q3URR9 | Gene names | Nudt17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 17, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 17). | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.172221 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.171891 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD17_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.214877 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C025 | Gene names | NUDT17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 17, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 17). | |||||
|
NUD13_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.218375 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUDT5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.121860 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |
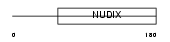
|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
AP4A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.110229 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50583 | Gene names | NUDT2, APAH1 | |||
|
Domain Architecture |
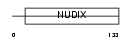
|
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
NUDT5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.118609 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKK9, Q6IAG0, Q9UH49 | Gene names | NUDT5 | |||
|
Domain Architecture |
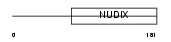
|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.017147 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AP4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.087560 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56380, Q9D2U6, Q9D6V2 | Gene names | Nudt2, Apah1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
REG1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.029596 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |
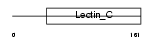
|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.028169 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |
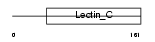
|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
NUD15_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.93392e-93 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD15_HUMAN
|
||||||
| NC score | 0.990451 (rank : 2) | θ value | 9.7205e-79 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
8ODP_MOUSE
|
||||||
| NC score | 0.431314 (rank : 3) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| NC score | 0.406858 (rank : 4) | θ value | 0.00020696 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
NUD13_MOUSE
|
||||||
| NC score | 0.229294 (rank : 5) | θ value | 0.00509761 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD17_MOUSE
|
||||||
| NC score | 0.225216 (rank : 6) | θ value | 0.00869519 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CWD3, Q3URR9 | Gene names | Nudt17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 17, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 17). | |||||
|
NUD13_HUMAN
|
||||||
| NC score | 0.218375 (rank : 7) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD17_HUMAN
|
||||||
| NC score | 0.214877 (rank : 8) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C025 | Gene names | NUDT17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 17, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 17). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.172221 (rank : 9) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_HUMAN
|
||||||
| NC score | 0.171891 (rank : 10) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUDT5_MOUSE
|
||||||
| NC score | 0.121860 (rank : 11) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
NUDT5_HUMAN
|
||||||
| NC score | 0.118609 (rank : 12) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKK9, Q6IAG0, Q9UH49 | Gene names | NUDT5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H). | |||||
|
AP4A_HUMAN
|
||||||
| NC score | 0.110229 (rank : 13) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50583 | Gene names | NUDT2, APAH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
AP4A_MOUSE
|
||||||
| NC score | 0.087560 (rank : 14) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56380, Q9D2U6, Q9D6V2 | Gene names | Nudt2, Apah1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
REG1A_HUMAN
|
||||||
| NC score | 0.029596 (rank : 15) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG1B_HUMAN
|
||||||
| NC score | 0.028169 (rank : 16) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.017147 (rank : 17) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||