Please be patient as the page loads
|
NUDT5_HUMAN
|
||||||
| SwissProt Accessions | Q9UKK9, Q6IAG0, Q9UH49 | Gene names | NUDT5 | |||
|
Domain Architecture |
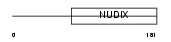
|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUDT5_HUMAN
|
||||||
| θ value | 2.79562e-118 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKK9, Q6IAG0, Q9UH49 | Gene names | NUDT5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H). | |||||
|
NUDT5_MOUSE
|
||||||
| θ value | 9.34977e-98 (rank : 2) | NC score | 0.995813 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |
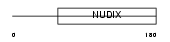
|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 0.47712 (rank : 3) | NC score | 0.090861 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.091711 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD15_HUMAN
|
||||||
| θ value | 1.38821 (rank : 5) | NC score | 0.110021 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUD15_MOUSE
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.118609 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUDT6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 7) | NC score | 0.050312 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH40, Q8R2T3 | Gene names | Nudt6, Asfgf2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Antisense basic fibroblast growth factor B). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.012259 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
NUDT5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.79562e-118 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKK9, Q6IAG0, Q9UH49 | Gene names | NUDT5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5) (YSA1H). | |||||
|
NUDT5_MOUSE
|
||||||
| NC score | 0.995813 (rank : 2) | θ value | 9.34977e-98 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKX6, Q99KM4 | Gene names | Nudt5 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-sugar pyrophosphatase (EC 3.6.1.13) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 5) (Nudix motif 5). | |||||
|
NUD15_MOUSE
|
||||||
| NC score | 0.118609 (rank : 3) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD15_HUMAN
|
||||||
| NC score | 0.110021 (rank : 4) | θ value | 1.38821 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUD12_HUMAN
|
||||||
| NC score | 0.091711 (rank : 5) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.090861 (rank : 6) | θ value | 0.47712 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUDT6_MOUSE
|
||||||
| NC score | 0.050312 (rank : 7) | θ value | 5.27518 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CH40, Q8R2T3 | Gene names | Nudt6, Asfgf2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Antisense basic fibroblast growth factor B). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.012259 (rank : 8) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||