Please be patient as the page loads
|
8ODP_MOUSE
|
||||||
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |
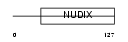
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
8ODP_MOUSE
|
||||||
| θ value | 9.74306e-71 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| θ value | 1.2784e-54 (rank : 2) | NC score | 0.985273 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |
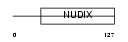
|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
NUD15_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.416412 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUD15_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.431314 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD12_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 5) | NC score | 0.248078 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 6) | NC score | 0.241899 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD13_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.284419 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD13_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.265720 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
MRP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.014413 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96J65, Q49AL2, Q8TAF0, Q8TEY2 | Gene names | ABCC12, MRP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 9 (ATP-binding cassette transporter sub-family C member 12). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.009096 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
8ODP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.74306e-71 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53368, P97795 | Gene names | Nudt1, Mth1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
8ODP_HUMAN
|
||||||
| NC score | 0.985273 (rank : 2) | θ value | 1.2784e-54 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36639, Q6P0Y6, Q9UBM0, Q9UBM9 | Gene names | NUDT1, MTH1 | |||
|
Domain Architecture |

|
|||||
| Description | 7,8-dihydro-8-oxoguanine triphosphatase (EC 3.1.6.-) (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1). | |||||
|
NUD15_MOUSE
|
||||||
| NC score | 0.431314 (rank : 3) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG93 | Gene names | Nudt15, Mth2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15) (MutT homolog 2) (mMTH2). | |||||
|
NUD15_HUMAN
|
||||||
| NC score | 0.416412 (rank : 4) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NV35, Q6P2C9 | Gene names | NUDT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable 7,8-dihydro-8-oxoguanine triphosphatase NUDT15 (EC 3.1.6.-) (8-oxo-dGTPase NUDT15) (Nucleoside diphosphate-linked moiety X motif 15) (Nudix motif 15). | |||||
|
NUD13_MOUSE
|
||||||
| NC score | 0.284419 (rank : 5) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZU0, Q9CXN4 | Gene names | Nudt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD13_HUMAN
|
||||||
| NC score | 0.265720 (rank : 6) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X67, Q5SQM4, Q5SQM5, Q9Y3X2 | Gene names | NUDT13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 13 (EC 3.-.-.-) (Nudix motif 13). | |||||
|
NUD12_HUMAN
|
||||||
| NC score | 0.248078 (rank : 7) | θ value | 0.000602161 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQG2, Q8TAL7 | Gene names | NUDT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.241899 (rank : 8) | θ value | 0.000602161 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
MRP9_HUMAN
|
||||||
| NC score | 0.014413 (rank : 9) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96J65, Q49AL2, Q8TAF0, Q8TEY2 | Gene names | ABCC12, MRP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 9 (ATP-binding cassette transporter sub-family C member 12). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.009096 (rank : 10) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||