Please be patient as the page loads
|
NPCL1_MOUSE
|
||||||
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.968164 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15118, Q9P130 | Gene names | NPC1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
NPC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.968062 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35604, O35605 | Gene names | Npc1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
NPCL1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996665 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHC9, Q6R3Q4, Q9UHC8 | Gene names | NPC1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
NPCL1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
PTC2_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 5) | NC score | 0.569033 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
PTC1_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 6) | NC score | 0.567093 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13635, Q13463, Q5VZC0 | Gene names | PTCH1, PTCH | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
PTC2_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 7) | NC score | 0.563846 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
PTC1_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 8) | NC score | 0.565977 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 9) | NC score | 0.292011 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
CY24B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.014013 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61093 | Gene names | Cybb, Cgd | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta). | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.014102 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
NBEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.014500 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.014407 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
SRTD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.018877 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.004810 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.007040 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.007399 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
ACE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.008218 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0I0, Q99N70, Q99N71 | Gene names | Ace2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.001471 (rank : 23) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CSF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.013689 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09919 | Gene names | CSF3 | |||
|
Domain Architecture |
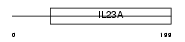
|
|||||
| Description | Granulocyte colony-stimulating factor precursor (G-CSF) (Pluripoietin) (Filgrastim) (Lenograstim). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.004544 (rank : 22) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
HMDH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.092076 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04035, Q8N190 | Gene names | HMGCR | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
HMDH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.091430 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01237, Q5U4I2 | Gene names | Hmgcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
NPCL1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6T3U4, Q5SVX1 | Gene names | Npc1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
NPCL1_HUMAN
|
||||||
| NC score | 0.996665 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHC9, Q6R3Q4, Q9UHC8 | Gene names | NPC1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niemann-Pick C1-like protein 1 precursor. | |||||
|
NPC1_HUMAN
|
||||||
| NC score | 0.968164 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15118, Q9P130 | Gene names | NPC1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
NPC1_MOUSE
|
||||||
| NC score | 0.968062 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35604, O35605 | Gene names | Npc1 | |||
|
Domain Architecture |

|
|||||
| Description | Niemann-Pick C1 protein precursor. | |||||
|
PTC2_MOUSE
|
||||||
| NC score | 0.569033 (rank : 5) | θ value | 3.88503e-19 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
PTC1_HUMAN
|
||||||
| NC score | 0.567093 (rank : 6) | θ value | 1.13037e-18 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13635, Q13463, Q5VZC0 | Gene names | PTCH1, PTCH | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
PTC1_MOUSE
|
||||||
| NC score | 0.565977 (rank : 7) | θ value | 3.28887e-18 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
PTC2_HUMAN
|
||||||
| NC score | 0.563846 (rank : 8) | θ value | 2.5182e-18 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.292011 (rank : 9) | θ value | 2.61198e-07 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
HMDH_HUMAN
|
||||||
| NC score | 0.092076 (rank : 10) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04035, Q8N190 | Gene names | HMGCR | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
HMDH_MOUSE
|
||||||
| NC score | 0.091430 (rank : 11) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01237, Q5U4I2 | Gene names | Hmgcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (EC 1.1.1.34) (HMG-CoA reductase). | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.018877 (rank : 12) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
NBEA_HUMAN
|
||||||
| NC score | 0.014500 (rank : 13) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.014407 (rank : 14) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.014102 (rank : 15) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
CY24B_MOUSE
|
||||||
| NC score | 0.014013 (rank : 16) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61093 | Gene names | Cybb, Cgd | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta). | |||||
|
CSF3_HUMAN
|
||||||
| NC score | 0.013689 (rank : 17) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09919 | Gene names | CSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Granulocyte colony-stimulating factor precursor (G-CSF) (Pluripoietin) (Filgrastim) (Lenograstim). | |||||
|
ACE2_MOUSE
|
||||||
| NC score | 0.008218 (rank : 18) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0I0, Q99N70, Q99N71 | Gene names | Ace2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.007399 (rank : 19) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.007040 (rank : 20) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.004810 (rank : 21) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.004544 (rank : 22) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.001471 (rank : 23) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||