Please be patient as the page loads
|
MA2A1_HUMAN
|
||||||
| SwissProt Accessions | Q16706, Q16767 | Gene names | MAN2A1, MANA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MA2A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16706, Q16767 | Gene names | MAN2A1, MANA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1). | |||||
|
MA2A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997220 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27046 | Gene names | Man2a1, Mana2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1) (AMAN II). | |||||
|
MA2A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.989476 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49641, Q13754 | Gene names | MAN2A2, MANA2X | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase IIx (EC 3.2.1.114) (Mannosyl-oligosaccharide 1,3- 1,6-alpha-mannosidase) (MAN IIx) (Mannosidase alpha class 2A member 2). | |||||
|
MA2B1_MOUSE
|
||||||
| θ value | 2.39424e-77 (rank : 4) | NC score | 0.873372 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09159, O55037, Q64443, Q9DBQ1 | Gene names | Man2b1, Laman, Man2b, Manb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase, alpha B) (Lysosomal acid alpha-mannosidase) (Laman) (Mannosidase alpha class 2B member 1). | |||||
|
MA2B2_MOUSE
|
||||||
| θ value | 6.96618e-77 (rank : 5) | NC score | 0.869904 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54782, Q69ZV1, Q8BH85, Q9DBK2 | Gene names | Man2b2, Kiaa0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
MA2B2_HUMAN
|
||||||
| θ value | 7.70199e-76 (rank : 6) | NC score | 0.872284 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2E5, Q66MP2, Q86T67 | Gene names | MAN2B2, KIAA0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
MA2B1_HUMAN
|
||||||
| θ value | 1.71583e-75 (rank : 7) | NC score | 0.870359 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00754, O15330, Q16680, Q93094, Q9BW13 | Gene names | MAN2B1, LAMAN, MANB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase, alpha B) (Lysosomal acid alpha-mannosidase) (Laman) (Mannosidase alpha class 2B member 1) [Contains: Lysosomal alpha-mannosidase A peptide; Lysosomal alpha-mannosidase B peptide; Lysosomal alpha-mannosidase C peptide; Lysosomal alpha-mannosidase D peptide; Lysosomal alpha- mannosidase E peptide]. | |||||
|
MA2C1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 8) | NC score | 0.297152 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91W89 | Gene names | Man2c1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1). | |||||
|
MA2C1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 9) | NC score | 0.258989 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTJ4, Q13358, Q9UL64 | Gene names | MAN2C1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1) (Alpha mannosidase 6A8B). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.006728 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.006684 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
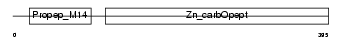
CBPA2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.029521 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.010796 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
MUC15_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.030813 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C6Z1, Q3UQC4, Q6XYQ9, Q7TMH6, Q8CE82 | Gene names | Muc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
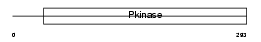
KCC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.003147 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.013451 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.013290 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
PCD10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.002204 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||
|
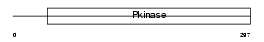
KCC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.003613 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16566 | Gene names | CAMK4 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.009512 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.014041 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.012089 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.014273 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.013007 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.007557 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
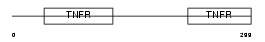
TNR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.008018 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
MA2A1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16706, Q16767 | Gene names | MAN2A1, MANA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1). | |||||
|
MA2A1_MOUSE
|
||||||
| NC score | 0.997220 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27046 | Gene names | Man2a1, Mana2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2 (EC 3.2.1.114) (Alpha-mannosidase II) (Mannosyl- oligosaccharide 1,3-1,6-alpha-mannosidase) (MAN II) (Golgi alpha- mannosidase II) (Mannosidase alpha class 2A member 1) (AMAN II). | |||||
|
MA2A2_HUMAN
|
||||||
| NC score | 0.989476 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49641, Q13754 | Gene names | MAN2A2, MANA2X | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase IIx (EC 3.2.1.114) (Mannosyl-oligosaccharide 1,3- 1,6-alpha-mannosidase) (MAN IIx) (Mannosidase alpha class 2A member 2). | |||||
|
MA2B1_MOUSE
|
||||||
| NC score | 0.873372 (rank : 4) | θ value | 2.39424e-77 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09159, O55037, Q64443, Q9DBQ1 | Gene names | Man2b1, Laman, Man2b, Manb | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase, alpha B) (Lysosomal acid alpha-mannosidase) (Laman) (Mannosidase alpha class 2B member 1). | |||||
|
MA2B2_HUMAN
|
||||||
| NC score | 0.872284 (rank : 5) | θ value | 7.70199e-76 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2E5, Q66MP2, Q86T67 | Gene names | MAN2B2, KIAA0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
MA2B1_HUMAN
|
||||||
| NC score | 0.870359 (rank : 6) | θ value | 1.71583e-75 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00754, O15330, Q16680, Q93094, Q9BW13 | Gene names | MAN2B1, LAMAN, MANB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase, alpha B) (Lysosomal acid alpha-mannosidase) (Laman) (Mannosidase alpha class 2B member 1) [Contains: Lysosomal alpha-mannosidase A peptide; Lysosomal alpha-mannosidase B peptide; Lysosomal alpha-mannosidase C peptide; Lysosomal alpha-mannosidase D peptide; Lysosomal alpha- mannosidase E peptide]. | |||||
|
MA2B2_MOUSE
|
||||||
| NC score | 0.869904 (rank : 7) | θ value | 6.96618e-77 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54782, Q69ZV1, Q8BH85, Q9DBK2 | Gene names | Man2b2, Kiaa0935 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymis-specific alpha-mannosidase precursor (EC 3.2.1.24) (Mannosidase alpha class 2B member 2). | |||||
|
MA2C1_MOUSE
|
||||||
| NC score | 0.297152 (rank : 8) | θ value | 8.40245e-06 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91W89 | Gene names | Man2c1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1). | |||||
|
MA2C1_HUMAN
|
||||||
| NC score | 0.258989 (rank : 9) | θ value | 7.1131e-05 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTJ4, Q13358, Q9UL64 | Gene names | MAN2C1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-mannosidase 2C1 (EC 3.2.1.24) (Alpha-D-mannoside mannohydrolase) (Mannosidase alpha class 2C member 1) (Alpha mannosidase 6A8B). | |||||
|
MUC15_MOUSE
|
||||||
| NC score | 0.030813 (rank : 10) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C6Z1, Q3UQC4, Q6XYQ9, Q7TMH6, Q8CE82 | Gene names | Muc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
CBPA2_HUMAN
|
||||||
| NC score | 0.029521 (rank : 11) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48052, Q96A12, Q96QN3 | Gene names | CPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase A2 precursor (EC 3.4.17.15). | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.014273 (rank : 12) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.014041 (rank : 13) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.013451 (rank : 14) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.013290 (rank : 15) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.013007 (rank : 16) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.012089 (rank : 17) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.010796 (rank : 18) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.009512 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.008018 (rank : 20) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.007557 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.006728 (rank : 22) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.006684 (rank : 23) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
KCC4_HUMAN
|
||||||
| NC score | 0.003613 (rank : 24) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16566 | Gene names | CAMK4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
KCC4_MOUSE
|
||||||
| NC score | 0.003147 (rank : 25) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
PCD10_HUMAN
|
||||||
| NC score | 0.002204 (rank : 26) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||