Please be patient as the page loads
|
LYAG_MOUSE
|
||||||
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LYAG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997908 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
LYAG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
MGA_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.961113 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
SUIS_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.961107 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
GANC_MOUSE
|
||||||
| θ value | 1.54472e-92 (rank : 5) | NC score | 0.905306 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
GANAB_MOUSE
|
||||||
| θ value | 1.70789e-91 (rank : 6) | NC score | 0.899157 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANC_HUMAN
|
||||||
| θ value | 7.17548e-90 (rank : 7) | NC score | 0.902986 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANAB_HUMAN
|
||||||
| θ value | 5.14231e-88 (rank : 8) | NC score | 0.899057 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
TFF3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.229763 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |
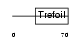
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 10) | NC score | 0.178581 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |
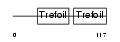
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.163680 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03404, O08913 | Gene names | Tff2, Sml1, Sp | |||
|
Domain Architecture |
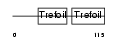
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP). | |||||
|
TFF1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.195523 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |
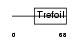
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.200449 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.201052 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
ZP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.034129 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.004695 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
VAPA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.013813 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
GI24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.013349 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H7M9, Q6UXF3, Q8WUG3, Q8WYZ8 | Gene names | C10orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet receptor Gi24 precursor. | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.002772 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
NADAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.012828 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
ZBTB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | -0.000685 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
ENKUR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.010556 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
FMOD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.001998 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06828, Q15331 | Gene names | FMOD, FM | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.002156 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.005536 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
LYAG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
LYAG_HUMAN
|
||||||
| NC score | 0.997908 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.961113 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
SUIS_HUMAN
|
||||||
| NC score | 0.961107 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
GANC_MOUSE
|
||||||
| NC score | 0.905306 (rank : 5) | θ value | 1.54472e-92 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
GANC_HUMAN
|
||||||
| NC score | 0.902986 (rank : 6) | θ value | 7.17548e-90 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANAB_MOUSE
|
||||||
| NC score | 0.899157 (rank : 7) | θ value | 1.70789e-91 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANAB_HUMAN
|
||||||
| NC score | 0.899057 (rank : 8) | θ value | 5.14231e-88 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
TFF3_MOUSE
|
||||||
| NC score | 0.229763 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF3_HUMAN
|
||||||
| NC score | 0.201052 (rank : 10) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
TFF1_MOUSE
|
||||||
| NC score | 0.200449 (rank : 11) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF1_HUMAN
|
||||||
| NC score | 0.195523 (rank : 12) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF2_HUMAN
|
||||||
| NC score | 0.178581 (rank : 13) | θ value | 0.00390308 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF2_MOUSE
|
||||||
| NC score | 0.163680 (rank : 14) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03404, O08913 | Gene names | Tff2, Sml1, Sp | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP). | |||||
|
ZP4_HUMAN
|
||||||
| NC score | 0.034129 (rank : 15) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
VAPA_MOUSE
|
||||||
| NC score | 0.013813 (rank : 16) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
GI24_HUMAN
|
||||||
| NC score | 0.013349 (rank : 17) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H7M9, Q6UXF3, Q8WUG3, Q8WYZ8 | Gene names | C10orf54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet receptor Gi24 precursor. | |||||
|
NADAP_HUMAN
|
||||||
| NC score | 0.012828 (rank : 18) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
ENKUR_MOUSE
|
||||||
| NC score | 0.010556 (rank : 19) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6SP97 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.005536 (rank : 20) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.004695 (rank : 21) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.002772 (rank : 22) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.002156 (rank : 23) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
FMOD_HUMAN
|
||||||
| NC score | 0.001998 (rank : 24) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06828, Q15331 | Gene names | FMOD, FM | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
ZBTB1_MOUSE
|
||||||
| NC score | -0.000685 (rank : 25) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||