Please be patient as the page loads
|
GANAB_MOUSE
|
||||||
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GANAB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997308 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
GANAB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANC_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987429 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANC_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987395 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
LYAG_MOUSE
|
||||||
| θ value | 1.70789e-91 (rank : 5) | NC score | 0.899157 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
LYAG_HUMAN
|
||||||
| θ value | 2.91321e-91 (rank : 6) | NC score | 0.899464 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
MGA_HUMAN
|
||||||
| θ value | 2.02684e-76 (rank : 7) | NC score | 0.863587 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
SUIS_HUMAN
|
||||||
| θ value | 3.57772e-73 (rank : 8) | NC score | 0.865740 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.028557 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.010240 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.089091 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.015786 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.035459 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
ZN646_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.002794 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
CLD23_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.021997 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |
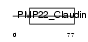
|
|||||
| Description | Claudin-23. | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.026936 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.021232 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.006656 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.007394 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.006869 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.009141 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
WBP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.014199 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.008914 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.005186 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
CAH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.008686 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.004543 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.010491 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.002710 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.005086 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | -0.001192 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
TFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.065607 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |
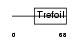
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.068867 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |
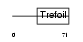
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.055080 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |
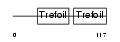
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.067804 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
TFF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.084119 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
GANAB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANAB_HUMAN
|
||||||
| NC score | 0.997308 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
GANC_HUMAN
|
||||||
| NC score | 0.987429 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANC_MOUSE
|
||||||
| NC score | 0.987395 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
LYAG_HUMAN
|
||||||
| NC score | 0.899464 (rank : 5) | θ value | 2.91321e-91 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
LYAG_MOUSE
|
||||||
| NC score | 0.899157 (rank : 6) | θ value | 1.70789e-91 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
SUIS_HUMAN
|
||||||
| NC score | 0.865740 (rank : 7) | θ value | 3.57772e-73 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.863587 (rank : 8) | θ value | 2.02684e-76 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.089091 (rank : 9) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TFF3_MOUSE
|
||||||
| NC score | 0.084119 (rank : 10) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF1_MOUSE
|
||||||
| NC score | 0.068867 (rank : 11) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF3_HUMAN
|
||||||
| NC score | 0.067804 (rank : 12) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
TFF1_HUMAN
|
||||||
| NC score | 0.065607 (rank : 13) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF2_HUMAN
|
||||||
| NC score | 0.055080 (rank : 14) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.035459 (rank : 15) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.028557 (rank : 16) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.026936 (rank : 17) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
CLD23_HUMAN
|
||||||
| NC score | 0.021997 (rank : 18) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.021232 (rank : 19) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.015786 (rank : 20) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
WBP4_HUMAN
|
||||||
| NC score | 0.014199 (rank : 21) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.010491 (rank : 22) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.010240 (rank : 23) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.009141 (rank : 24) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.008914 (rank : 25) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CAH9_HUMAN
|
||||||
| NC score | 0.008686 (rank : 26) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.007394 (rank : 27) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.006869 (rank : 28) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.006656 (rank : 29) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.005186 (rank : 30) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.005086 (rank : 31) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.004543 (rank : 32) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ZN646_HUMAN
|
||||||
| NC score | 0.002794 (rank : 33) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.002710 (rank : 34) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | -0.001192 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||