Please be patient as the page loads
|
TFF2_MOUSE
|
||||||
| SwissProt Accessions | Q03404, O08913 | Gene names | Tff2, Sml1, Sp | |||
|
Domain Architecture |
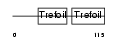
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TFF2_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03404, O08913 | Gene names | Tff2, Sml1, Sp | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP). | |||||
|
TFF2_HUMAN
|
||||||
| θ value | 7.24236e-58 (rank : 2) | NC score | 0.992314 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |
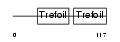
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF1_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 3) | NC score | 0.833896 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |
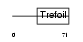
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 4) | NC score | 0.819130 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |
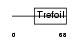
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF3_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 5) | NC score | 0.801739 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF3_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 6) | NC score | 0.777910 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
LYAG_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.163680 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
LYAG_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.158367 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
KR101_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.062918 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.061546 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR414_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.048525 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.051892 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.053334 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.031797 (rank : 16) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
MGA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.066093 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
SUIS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.060966 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
TFF2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.24614e-67 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03404, O08913 | Gene names | Tff2, Sml1, Sp | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP). | |||||
|
TFF2_HUMAN
|
||||||
| NC score | 0.992314 (rank : 2) | θ value | 7.24236e-58 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF1_MOUSE
|
||||||
| NC score | 0.833896 (rank : 3) | θ value | 1.86753e-13 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF1_HUMAN
|
||||||
| NC score | 0.819130 (rank : 4) | θ value | 3.52202e-12 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF3_MOUSE
|
||||||
| NC score | 0.801739 (rank : 5) | θ value | 3.89403e-11 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF3_HUMAN
|
||||||
| NC score | 0.777910 (rank : 6) | θ value | 1.80886e-08 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
LYAG_MOUSE
|
||||||
| NC score | 0.163680 (rank : 7) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
LYAG_HUMAN
|
||||||
| NC score | 0.158367 (rank : 8) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.066093 (rank : 9) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.062918 (rank : 10) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.061546 (rank : 11) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
SUIS_HUMAN
|
||||||
| NC score | 0.060966 (rank : 12) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.053334 (rank : 13) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.051892 (rank : 14) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.048525 (rank : 15) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.031797 (rank : 16) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||