Please be patient as the page loads
|
C1TC_HUMAN
|
||||||
| SwissProt Accessions | P11586, Q86VC9, Q9BVP5 | Gene names | MTHFD1, MTHFC, MTHFD | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
C1TC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11586, Q86VC9, Q9BVP5 | Gene names | MTHFD1, MTHFC, MTHFD | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
C1TC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998363 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q922D8 | Gene names | Mthfd1 | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
MTDC_MOUSE
|
||||||
| θ value | 7.75577e-52 (rank : 3) | NC score | 0.844634 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18155, Q3TMN4 | Gene names | Mthfd2, Nmdmc | |||
|
Domain Architecture |
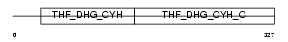
|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
MTDC_HUMAN
|
||||||
| θ value | 1.11993e-50 (rank : 4) | NC score | 0.842072 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13995, Q53G90, Q53GV5, Q53S36 | Gene names | MTHFD2, NMDMC | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | 1.38821 (rank : 5) | NC score | 0.014709 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.016652 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CR034_MOUSE
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.011420 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
TRI10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.010415 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UDY6, Q86Z08, Q96QB6, Q9C023, Q9C024 | Gene names | TRIM10, RFB30, RNF9 | |||
|
Domain Architecture |
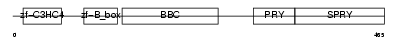
|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (B30- RING finger protein). | |||||
|
CRYM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.021284 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54983 | Gene names | Crym | |||
|
Domain Architecture |
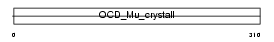
|
|||||
| Description | Mu-crystallin homolog. | |||||
|
NNTM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.026272 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
YMEL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.011656 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.011444 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.004510 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
ITAL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.010699 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.013574 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.013582 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
RGMC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.011792 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQ32, Q8CEU7, Q8K1D4, Q9D741 | Gene names | Hfe2, Rgmc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemojuvelin precursor (Hemochromatosis type 2 protein homolog) (RGM domain family member C). | |||||
|
EDEM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.008617 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.001593 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
C1TC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11586, Q86VC9, Q9BVP5 | Gene names | MTHFD1, MTHFC, MTHFD | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
C1TC_MOUSE
|
||||||
| NC score | 0.998363 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q922D8 | Gene names | Mthfd1 | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
MTDC_MOUSE
|
||||||
| NC score | 0.844634 (rank : 3) | θ value | 7.75577e-52 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18155, Q3TMN4 | Gene names | Mthfd2, Nmdmc | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
MTDC_HUMAN
|
||||||
| NC score | 0.842072 (rank : 4) | θ value | 1.11993e-50 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13995, Q53G90, Q53GV5, Q53S36 | Gene names | MTHFD2, NMDMC | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial precursor [Includes: NAD-dependent methylenetetrahydrofolate dehydrogenase (EC 1.5.1.15); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9)]. | |||||
|
NNTM_MOUSE
|
||||||
| NC score | 0.026272 (rank : 5) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
CRYM_MOUSE
|
||||||
| NC score | 0.021284 (rank : 6) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54983 | Gene names | Crym | |||
|
Domain Architecture |
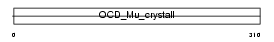
|
|||||
| Description | Mu-crystallin homolog. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.016652 (rank : 7) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.014709 (rank : 8) | θ value | 1.38821 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.013582 (rank : 9) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.013574 (rank : 10) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
RGMC_MOUSE
|
||||||
| NC score | 0.011792 (rank : 11) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQ32, Q8CEU7, Q8K1D4, Q9D741 | Gene names | Hfe2, Rgmc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemojuvelin precursor (Hemochromatosis type 2 protein homolog) (RGM domain family member C). | |||||
|
YMEL1_HUMAN
|
||||||
| NC score | 0.011656 (rank : 12) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96TA2, Q9H1Q0, Q9UMR9 | Gene names | YME1L1, FTSH1, YME1L | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1) (Meg-4) (Presenilin- associated metalloprotease) (PAMP). | |||||
|
YMEL1_MOUSE
|
||||||
| NC score | 0.011444 (rank : 13) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88967, Q7TNN5 | Gene names | Yme1l1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent metalloprotease YME1L1 (EC 3.4.24.-) (YME1-like protein 1) (ATP-dependent metalloprotease FtsH1). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.011420 (rank : 14) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
ITAL_MOUSE
|
||||||
| NC score | 0.010699 (rank : 15) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
TRI10_HUMAN
|
||||||
| NC score | 0.010415 (rank : 16) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UDY6, Q86Z08, Q96QB6, Q9C023, Q9C024 | Gene names | TRIM10, RFB30, RNF9 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 10 (RING finger protein 9) (B30- RING finger protein). | |||||
|
EDEM1_HUMAN
|
||||||
| NC score | 0.008617 (rank : 17) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.004510 (rank : 18) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.001593 (rank : 19) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||