Please be patient as the page loads
|
ATP9B_HUMAN
|
||||||
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATP9A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997365 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
ATP9A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997640 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
ATP9B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998638 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
AT11B_HUMAN
|
||||||
| θ value | 3.5201e-129 (rank : 5) | NC score | 0.955702 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 4.02752e-125 (rank : 6) | NC score | 0.945593 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT8A1_MOUSE
|
||||||
| θ value | 3.76964e-123 (rank : 7) | NC score | 0.951002 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| θ value | 4.92331e-123 (rank : 8) | NC score | 0.950899 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT11A_HUMAN
|
||||||
| θ value | 2.06847e-121 (rank : 9) | NC score | 0.946247 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 9.60843e-119 (rank : 10) | NC score | 0.949316 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT8A2_HUMAN
|
||||||
| θ value | 2.79562e-118 (rank : 11) | NC score | 0.947586 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8B2_HUMAN
|
||||||
| θ value | 2.79562e-118 (rank : 12) | NC score | 0.947248 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT8A2_MOUSE
|
||||||
| θ value | 1.17455e-116 (rank : 13) | NC score | 0.948159 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT11C_HUMAN
|
||||||
| θ value | 1.09934e-114 (rank : 14) | NC score | 0.948163 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT8B1_HUMAN
|
||||||
| θ value | 6.6695e-112 (rank : 15) | NC score | 0.943678 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 2.73936e-73 (rank : 16) | NC score | 0.909303 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 1.7756e-72 (rank : 17) | NC score | 0.925971 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
AT10D_MOUSE
|
||||||
| θ value | 2.65329e-68 (rank : 18) | NC score | 0.905729 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_HUMAN
|
||||||
| θ value | 4.23606e-66 (rank : 19) | NC score | 0.903601 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 1.66577e-62 (rank : 20) | NC score | 0.900339 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10A_HUMAN
|
||||||
| θ value | 3.14151e-61 (rank : 21) | NC score | 0.897906 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT1A4_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 22) | NC score | 0.452978 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 23) | NC score | 0.596594 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT1A2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 24) | NC score | 0.449512 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 25) | NC score | 0.449008 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT132_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 26) | NC score | 0.588806 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT133_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 27) | NC score | 0.629719 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 28) | NC score | 0.461755 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2A1_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 29) | NC score | 0.454284 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 30) | NC score | 0.459522 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2C1_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 31) | NC score | 0.456734 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 32) | NC score | 0.454175 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
ATP4A_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 33) | NC score | 0.459713 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT131_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 34) | NC score | 0.530111 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT1A4_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 35) | NC score | 0.445445 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
ATP4A_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 36) | NC score | 0.455335 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2A1_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 37) | NC score | 0.450823 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 38) | NC score | 0.465824 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 39) | NC score | 0.463426 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 40) | NC score | 0.539355 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 41) | NC score | 0.539374 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT1A1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 42) | NC score | 0.440740 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 43) | NC score | 0.542523 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 44) | NC score | 0.421018 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 45) | NC score | 0.419018 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2B4_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 46) | NC score | 0.557615 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT1A1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 47) | NC score | 0.437465 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT131_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 48) | NC score | 0.523233 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT12A_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 49) | NC score | 0.457598 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 50) | NC score | 0.404834 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 51) | NC score | 0.458661 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 52) | NC score | 0.510834 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 53) | NC score | 0.005208 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.032629 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.002681 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
HNRPD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.007019 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |
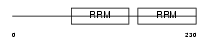
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.007037 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |
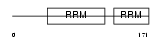
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.010856 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
RON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | -0.003770 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.002023 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | -0.000787 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.024202 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
PPIE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.003709 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |
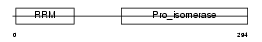
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.003696 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |
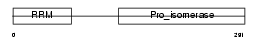
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.000706 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TENN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.000133 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
CA101_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.012810 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SY80, Q8IYZ6 | Gene names | C1orf101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf101 precursor. | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.000683 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | -0.001256 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PA24D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.002814 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
S26A6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007618 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXS9, Q96Q90 | Gene names | SLC26A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 26 member 6 (Pendrin-like protein 1) (Pendrin L1). | |||||
|
ATP7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.170358 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.168526 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
ATP7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.174083 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.172828 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
ATP9B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O43861, O60872 | Gene names | ATP9B, ATPIIB, NEO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9B_MOUSE
|
||||||
| NC score | 0.998638 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
ATP9A_MOUSE
|
||||||
| NC score | 0.997640 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O70228, Q8VDI5, Q922L9 | Gene names | Atp9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A). | |||||
|
ATP9A_HUMAN
|
||||||
| NC score | 0.997365 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75110, Q5TFW5, Q5TFW6, Q5TFW9, Q6ZMF3, Q9NQK6, Q9NQK7 | Gene names | ATP9A, ATPIIA, KIAA0611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IIA (EC 3.6.3.1) (ATPase class II type 9A) (ATPase IIA). | |||||
|
AT11B_HUMAN
|
||||||
| NC score | 0.955702 (rank : 5) | θ value | 3.5201e-129 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2G3, Q96FN1, Q9UKK7 | Gene names | ATP11B, ATPIF, ATPIR, KIAA0956 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IF (EC 3.6.3.1) (ATPase class I type 11B) (ATPase IR). | |||||
|
AT8A1_MOUSE
|
||||||
| NC score | 0.951002 (rank : 6) | θ value | 3.76964e-123 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_HUMAN
|
||||||
| NC score | 0.950899 (rank : 7) | θ value | 4.92331e-123 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.949316 (rank : 8) | θ value | 9.60843e-119 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
AT11C_HUMAN
|
||||||
| NC score | 0.948163 (rank : 9) | θ value | 1.09934e-114 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NB49, Q5JT69, Q5JT70, Q5JT71, Q5JT72, Q5JT73, Q6ZND5, Q6ZU50, Q6ZUP7, Q70IJ9, Q70IK0, Q8WX24 | Gene names | ATP11C, ATPIG, ATPIQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IG (EC 3.6.3.1) (ATPase class I type 11C) (ATPase IG) (ATPase IQ) (ATPase class VI type 11C). | |||||
|
AT8A2_MOUSE
|
||||||
| NC score | 0.948159 (rank : 10) | θ value | 1.17455e-116 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98200 | Gene names | Atp8a2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2). | |||||
|
AT8A2_HUMAN
|
||||||
| NC score | 0.947586 (rank : 11) | θ value | 2.79562e-118 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NTI2, Q9H527, Q9NPU6, Q9NTL2, Q9NYM3 | Gene names | ATP8A2, ATPIB | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1). | |||||
|
AT8B2_HUMAN
|
||||||
| NC score | 0.947248 (rank : 12) | θ value | 2.79562e-118 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98198, Q96I43, Q96NQ7 | Gene names | ATP8B2, ATPID, KIAA1137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase ID (EC 3.6.3.1) (ATPase class I type 8B member 2). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.946247 (rank : 13) | θ value | 2.06847e-121 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.945593 (rank : 14) | θ value | 4.02752e-125 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
AT8B1_HUMAN
|
||||||
| NC score | 0.943678 (rank : 15) | θ value | 6.6695e-112 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43520, Q9BTP8 | Gene names | ATP8B1, ATPIC, FIC1, PFIC | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IC (EC 3.6.3.1) (Familial intrahepatic cholestasis type 1) (ATPase class I type 8B member 1). | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.925971 (rank : 16) | θ value | 1.7756e-72 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.909303 (rank : 17) | θ value | 2.73936e-73 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT10D_MOUSE
|
||||||
| NC score | 0.905729 (rank : 18) | θ value | 2.65329e-68 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2X1 | Gene names | Atp10d | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10D_HUMAN
|
||||||
| NC score | 0.903601 (rank : 19) | θ value | 4.23606e-66 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.900339 (rank : 20) | θ value | 1.66577e-62 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.897906 (rank : 21) | θ value | 3.14151e-61 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
AT133_HUMAN
|
||||||
| NC score | 0.629719 (rank : 22) | θ value | 1.74796e-11 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H7F0, Q8NC11, Q96KS1 | Gene names | ATP13A3, AFURS1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A3 (EC 3.6.3.-) (ATPase family homolog up-regulated in senescence cells 1). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.596594 (rank : 23) | θ value | 4.44505e-15 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT132_HUMAN
|
||||||
| NC score | 0.588806 (rank : 24) | θ value | 2.43908e-13 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
AT2B4_HUMAN
|
||||||
| NC score | 0.557615 (rank : 25) | θ value | 5.26297e-08 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.542523 (rank : 26) | θ value | 4.0297e-08 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.539374 (rank : 27) | θ value | 2.36244e-08 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.539355 (rank : 28) | θ value | 1.80886e-08 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT131_MOUSE
|
||||||
| NC score | 0.530111 (rank : 29) | θ value | 3.64472e-09 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPE9 | Gene names | Atp13a1, Atp13a | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-) (CATP). | |||||
|
AT131_HUMAN
|
||||||
| NC score | 0.523233 (rank : 30) | θ value | 1.53129e-07 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HD20, Q9H6C6 | Gene names | ATP13A1, ATP13A, KIAA1825 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A1 (EC 3.6.3.-). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.510834 (rank : 31) | θ value | 8.40245e-06 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT2A2_MOUSE
|
||||||
| NC score | 0.465824 (rank : 32) | θ value | 4.76016e-09 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_HUMAN
|
||||||
| NC score | 0.463426 (rank : 33) | θ value | 8.11959e-09 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.461755 (rank : 34) | θ value | 1.74796e-11 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
ATP4A_MOUSE
|
||||||
| NC score | 0.459713 (rank : 35) | θ value | 2.79066e-09 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64436, Q9CV46 | Gene names | Atp4a | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2A3_HUMAN
|
||||||
| NC score | 0.459522 (rank : 36) | θ value | 9.59137e-10 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.458661 (rank : 37) | θ value | 2.21117e-06 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT12A_MOUSE
|
||||||
| NC score | 0.457598 (rank : 38) | θ value | 3.41135e-07 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z1W8, Q8VHY2 | Gene names | Atp12a, Atp1al1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
AT2C1_MOUSE
|
||||||
| NC score | 0.456734 (rank : 39) | θ value | 9.59137e-10 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
ATP4A_HUMAN
|
||||||
| NC score | 0.455335 (rank : 40) | θ value | 3.64472e-09 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20648, O00738 | Gene names | ATP4A | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 1 (EC 3.6.3.10) (Proton pump) (Gastric H+/K+ ATPase subunit alpha). | |||||
|
AT2A1_MOUSE
|
||||||
| NC score | 0.454284 (rank : 41) | θ value | 2.52405e-10 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.454175 (rank : 42) | θ value | 1.25267e-09 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT1A4_HUMAN
|
||||||
| NC score | 0.452978 (rank : 43) | θ value | 1.38178e-16 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13733, Q504T2, Q8TBN8, Q8WXA7, Q8WXH7, Q8WY13 | Gene names | ATP1A4, ATP1AL2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT2A1_HUMAN
|
||||||
| NC score | 0.450823 (rank : 44) | θ value | 4.76016e-09 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT1A2_HUMAN
|
||||||
| NC score | 0.449512 (rank : 45) | θ value | 4.91457e-14 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P50993, Q07059, Q86UZ5, Q9UQ25 | Gene names | ATP1A2, KIAA0778 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2). | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.449008 (rank : 46) | θ value | 4.91457e-14 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
AT1A4_MOUSE
|
||||||
| NC score | 0.445445 (rank : 47) | θ value | 3.64472e-09 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WV27, Q9R173, Q9WV28 | Gene names | Atp1a4, Atp1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-4 chain (EC 3.6.3.9) (Sodium pump 4) (Na+/K+ ATPase 4). | |||||
|
AT1A1_MOUSE
|
||||||
| NC score | 0.440740 (rank : 48) | θ value | 3.08544e-08 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDN2, Q91Z09 | Gene names | Atp1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A1_HUMAN
|
||||||
| NC score | 0.437465 (rank : 49) | θ value | 8.97725e-08 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P05023, Q16689, Q6LDM4, Q9UJ20, Q9UJ21 | Gene names | ATP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-1 chain precursor (EC 3.6.3.9) (Sodium pump 1) (Na+/K+ ATPase 1). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.421018 (rank : 50) | θ value | 5.26297e-08 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.419018 (rank : 51) | θ value | 5.26297e-08 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.404834 (rank : 52) | θ value | 4.45536e-07 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
ATP7B_HUMAN
|
||||||
| NC score | 0.174083 (rank : 53) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35670, Q16318, Q16319, Q4U3V3, Q59FJ9, Q5T7X7 | Gene names | ATP7B, PWD, WC1, WND | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein). | |||||
|
ATP7B_MOUSE
|
||||||
| NC score | 0.172828 (rank : 54) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64446 | Gene names | Atp7b, Wnd | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 2 (EC 3.6.3.4) (Copper pump 2) (Wilson disease-associated protein homolog). | |||||
|
ATP7A_HUMAN
|
||||||
| NC score | 0.170358 (rank : 55) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04656, O00227, O00745, Q9BYY8 | Gene names | ATP7A, MC1, MNK | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein). | |||||
|
ATP7A_MOUSE
|
||||||
| NC score | 0.168526 (rank : 56) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64430, O35101, P97422, Q64431 | Gene names | Atp7a, Mnk | |||
|
Domain Architecture |

|
|||||
| Description | Copper-transporting ATPase 1 (EC 3.6.3.4) (Copper pump 1) (Menkes disease-associated protein homolog). | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.032629 (rank : 57) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.024202 (rank : 58) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CA101_HUMAN
|
||||||
| NC score | 0.012810 (rank : 59) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SY80, Q8IYZ6 | Gene names | C1orf101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf101 precursor. | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.010856 (rank : 60) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
S26A6_HUMAN
|
||||||
| NC score | 0.007618 (rank : 61) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXS9, Q96Q90 | Gene names | SLC26A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 26 member 6 (Pendrin-like protein 1) (Pendrin L1). | |||||
|
HNRPD_MOUSE
|
||||||
| NC score | 0.007037 (rank : 62) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_HUMAN
|
||||||
| NC score | 0.007019 (rank : 63) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.005208 (rank : 64) | θ value | 0.0563607 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
PPIE_HUMAN
|
||||||
| NC score | 0.003709 (rank : 65) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| NC score | 0.003696 (rank : 66) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PA24D_HUMAN
|
||||||
| NC score | 0.002814 (rank : 67) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.002681 (rank : 68) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.002023 (rank : 69) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.000706 (rank : 70) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.000683 (rank : 71) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
TENN_HUMAN
|
||||||
| NC score | 0.000133 (rank : 72) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
CP250_MOUSE
|
||||||
| NC score | -0.000787 (rank : 73) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NIN_HUMAN
|
||||||
| NC score | -0.001256 (rank : 74) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
RON_HUMAN
|
||||||
| NC score | -0.003770 (rank : 75) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||