Please be patient as the page loads
|
ACAD9_HUMAN
|
||||||
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
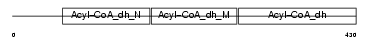
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACAD9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
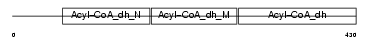
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACAD9_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999199 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
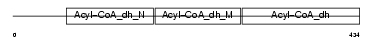
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADV_MOUSE
|
||||||
| θ value | 4.42207e-148 (rank : 3) | NC score | 0.986173 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
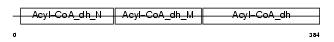
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACADV_HUMAN
|
||||||
| θ value | 2.77624e-142 (rank : 4) | NC score | 0.986462 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
ACDSB_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 5) | NC score | 0.944655 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACADS_HUMAN
|
||||||
| θ value | 7.24236e-58 (rank : 6) | NC score | 0.945950 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
IVD_MOUSE
|
||||||
| θ value | 1.61343e-57 (rank : 7) | NC score | 0.939248 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
IVD_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 8) | NC score | 0.937138 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACDSB_MOUSE
|
||||||
| θ value | 3.0428e-56 (rank : 9) | NC score | 0.939602 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACADS_MOUSE
|
||||||
| θ value | 5.19021e-56 (rank : 10) | NC score | 0.943545 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACADM_HUMAN
|
||||||
| θ value | 2.18062e-54 (rank : 11) | NC score | 0.937814 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADM_MOUSE
|
||||||
| θ value | 5.37103e-53 (rank : 12) | NC score | 0.936491 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACAD8_MOUSE
|
||||||
| θ value | 5.55816e-50 (rank : 13) | NC score | 0.929634 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
ACAD8_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 14) | NC score | 0.926453 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ACADL_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 15) | NC score | 0.918292 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
ACADL_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 16) | NC score | 0.914512 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
GCDH_HUMAN
|
||||||
| θ value | 1.24399e-33 (rank : 17) | NC score | 0.897474 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
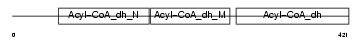
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
GCDH_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 18) | NC score | 0.891661 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
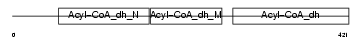
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACOX3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 19) | NC score | 0.468139 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 20) | NC score | 0.449613 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.306921 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 22) | NC score | 0.309317 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 23) | NC score | 0.242796 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.203917 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ZN317_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | -0.001380 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |
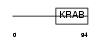
|
|||||
| Description | Zinc finger protein 317. | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.004440 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.004357 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.007622 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
ACAD9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
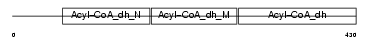
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACAD9_MOUSE
|
||||||
| NC score | 0.999199 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
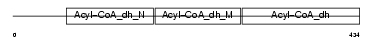
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADV_HUMAN
|
||||||
| NC score | 0.986462 (rank : 3) | θ value | 2.77624e-142 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |

|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
ACADV_MOUSE
|
||||||
| NC score | 0.986173 (rank : 4) | θ value | 4.42207e-148 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
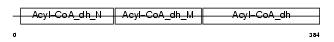
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACADS_HUMAN
|
||||||
| NC score | 0.945950 (rank : 5) | θ value | 7.24236e-58 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
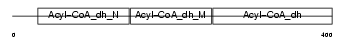
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACDSB_HUMAN
|
||||||
| NC score | 0.944655 (rank : 6) | θ value | 4.53632e-60 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
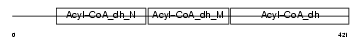
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACADS_MOUSE
|
||||||
| NC score | 0.943545 (rank : 7) | θ value | 5.19021e-56 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
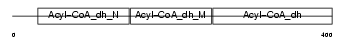
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACDSB_MOUSE
|
||||||
| NC score | 0.939602 (rank : 8) | θ value | 3.0428e-56 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
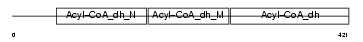
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
IVD_MOUSE
|
||||||
| NC score | 0.939248 (rank : 9) | θ value | 1.61343e-57 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACADM_HUMAN
|
||||||
| NC score | 0.937814 (rank : 10) | θ value | 2.18062e-54 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
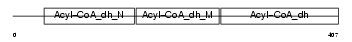
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
IVD_HUMAN
|
||||||
| NC score | 0.937138 (rank : 11) | θ value | 2.10721e-57 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
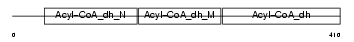
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACADM_MOUSE
|
||||||
| NC score | 0.936491 (rank : 12) | θ value | 5.37103e-53 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
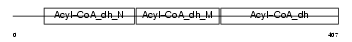
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACAD8_MOUSE
|
||||||
| NC score | 0.929634 (rank : 13) | θ value | 5.55816e-50 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
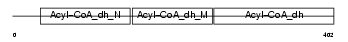
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
ACAD8_HUMAN
|
||||||
| NC score | 0.926453 (rank : 14) | θ value | 3.72821e-46 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
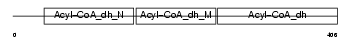
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ACADL_MOUSE
|
||||||
| NC score | 0.918292 (rank : 15) | θ value | 2.11701e-41 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
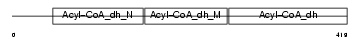
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
ACADL_HUMAN
|
||||||
| NC score | 0.914512 (rank : 16) | θ value | 8.04462e-41 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
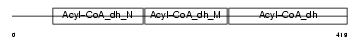
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
GCDH_HUMAN
|
||||||
| NC score | 0.897474 (rank : 17) | θ value | 1.24399e-33 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
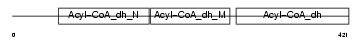
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
GCDH_MOUSE
|
||||||
| NC score | 0.891661 (rank : 18) | θ value | 1.6813e-30 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
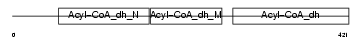
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACOX3_MOUSE
|
||||||
| NC score | 0.468139 (rank : 19) | θ value | 3.29651e-10 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_HUMAN
|
||||||
| NC score | 0.449613 (rank : 20) | θ value | 1.38499e-08 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_MOUSE
|
||||||
| NC score | 0.309317 (rank : 21) | θ value | 3.19293e-05 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX1_HUMAN
|
||||||
| NC score | 0.306921 (rank : 22) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX2_HUMAN
|
||||||
| NC score | 0.242796 (rank : 23) | θ value | 0.0193708 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_MOUSE
|
||||||
| NC score | 0.203917 (rank : 24) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.007622 (rank : 25) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.004440 (rank : 26) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.004357 (rank : 27) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
ZN317_HUMAN
|
||||||
| NC score | -0.001380 (rank : 28) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 317. | |||||