Please be patient as the page loads
|
ACADV_HUMAN
|
||||||
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |
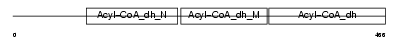
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACADV_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |

|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
ACADV_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998867 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
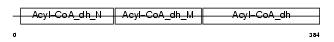
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACAD9_MOUSE
|
||||||
| θ value | 1.73892e-144 (rank : 3) | NC score | 0.986325 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
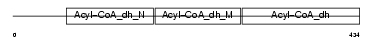
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACAD9_HUMAN
|
||||||
| θ value | 2.77624e-142 (rank : 4) | NC score | 0.986462 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
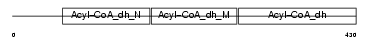
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
IVD_MOUSE
|
||||||
| θ value | 2.84797e-54 (rank : 5) | NC score | 0.935463 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACADS_HUMAN
|
||||||
| θ value | 3.71957e-54 (rank : 6) | NC score | 0.941692 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
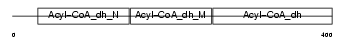
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACADS_MOUSE
|
||||||
| θ value | 2.41096e-53 (rank : 7) | NC score | 0.939444 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
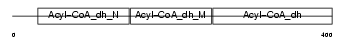
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACDSB_HUMAN
|
||||||
| θ value | 3.1488e-53 (rank : 8) | NC score | 0.939510 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
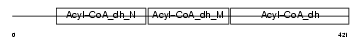
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
IVD_HUMAN
|
||||||
| θ value | 7.01481e-53 (rank : 9) | NC score | 0.933224 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
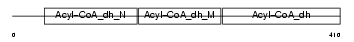
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACDSB_MOUSE
|
||||||
| θ value | 9.16162e-53 (rank : 10) | NC score | 0.935541 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
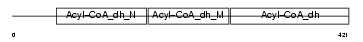
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACAD8_MOUSE
|
||||||
| θ value | 2.3352e-48 (rank : 11) | NC score | 0.927474 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
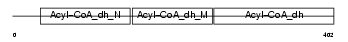
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
ACAD8_HUMAN
|
||||||
| θ value | 3.98324e-48 (rank : 12) | NC score | 0.925541 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
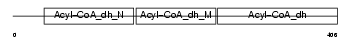
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ACADM_MOUSE
|
||||||
| θ value | 1.67352e-46 (rank : 13) | NC score | 0.929488 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
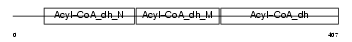
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADM_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 14) | NC score | 0.930469 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADL_MOUSE
|
||||||
| θ value | 9.50281e-42 (rank : 15) | NC score | 0.915639 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
ACADL_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 16) | NC score | 0.910555 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
GCDH_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 17) | NC score | 0.900456 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
GCDH_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 18) | NC score | 0.895289 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
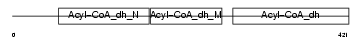
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACOX3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 19) | NC score | 0.441780 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 20) | NC score | 0.457297 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 21) | NC score | 0.302727 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 22) | NC score | 0.297973 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.236963 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.196512 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.005732 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.011463 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.009464 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
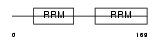
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
LMIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.014021 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
KLF10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | -0.001771 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
RAIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.009098 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
KS6A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | -0.003090 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75582, O95316, Q96AF7 | Gene names | RPS6KA5, MSK1 | |||
|
Domain Architecture |
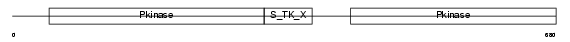
|
|||||
| Description | Ribosomal protein S6 kinase alpha-5 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 1) (90 kDa ribosomal protein S6 kinase 5) (RSK-like protein kinase) (RSKL). | |||||
|
ZN317_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | -0.001567 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |
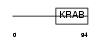
|
|||||
| Description | Zinc finger protein 317. | |||||
|
CTR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.008550 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
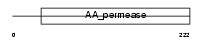
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.001017 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.006829 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
ACADV_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |

|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
ACADV_MOUSE
|
||||||
| NC score | 0.998867 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
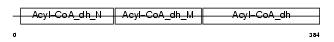
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACAD9_HUMAN
|
||||||
| NC score | 0.986462 (rank : 3) | θ value | 2.77624e-142 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
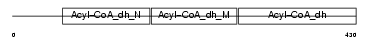
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACAD9_MOUSE
|
||||||
| NC score | 0.986325 (rank : 4) | θ value | 1.73892e-144 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
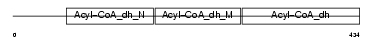
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADS_HUMAN
|
||||||
| NC score | 0.941692 (rank : 5) | θ value | 3.71957e-54 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACDSB_HUMAN
|
||||||
| NC score | 0.939510 (rank : 6) | θ value | 3.1488e-53 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACADS_MOUSE
|
||||||
| NC score | 0.939444 (rank : 7) | θ value | 2.41096e-53 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACDSB_MOUSE
|
||||||
| NC score | 0.935541 (rank : 8) | θ value | 9.16162e-53 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
IVD_MOUSE
|
||||||
| NC score | 0.935463 (rank : 9) | θ value | 2.84797e-54 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
IVD_HUMAN
|
||||||
| NC score | 0.933224 (rank : 10) | θ value | 7.01481e-53 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
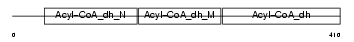
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACADM_HUMAN
|
||||||
| NC score | 0.930469 (rank : 11) | θ value | 8.30557e-46 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
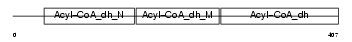
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADM_MOUSE
|
||||||
| NC score | 0.929488 (rank : 12) | θ value | 1.67352e-46 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
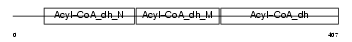
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACAD8_MOUSE
|
||||||
| NC score | 0.927474 (rank : 13) | θ value | 2.3352e-48 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
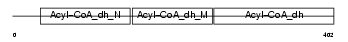
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
ACAD8_HUMAN
|
||||||
| NC score | 0.925541 (rank : 14) | θ value | 3.98324e-48 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
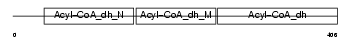
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ACADL_MOUSE
|
||||||
| NC score | 0.915639 (rank : 15) | θ value | 9.50281e-42 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
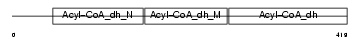
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
ACADL_HUMAN
|
||||||
| NC score | 0.910555 (rank : 16) | θ value | 4.88048e-38 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
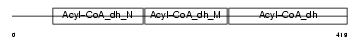
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
GCDH_HUMAN
|
||||||
| NC score | 0.900456 (rank : 17) | θ value | 1.32909e-35 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
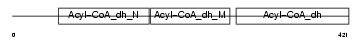
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
GCDH_MOUSE
|
||||||
| NC score | 0.895289 (rank : 18) | θ value | 2.12192e-33 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACOX3_MOUSE
|
||||||
| NC score | 0.457297 (rank : 19) | θ value | 4.0297e-08 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_HUMAN
|
||||||
| NC score | 0.441780 (rank : 20) | θ value | 4.0297e-08 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_MOUSE
|
||||||
| NC score | 0.302727 (rank : 21) | θ value | 1.43324e-05 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX1_HUMAN
|
||||||
| NC score | 0.297973 (rank : 22) | θ value | 0.00020696 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX2_HUMAN
|
||||||
| NC score | 0.236963 (rank : 23) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_MOUSE
|
||||||
| NC score | 0.196512 (rank : 24) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
LMIP_HUMAN
|
||||||
| NC score | 0.014021 (rank : 25) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.011463 (rank : 26) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.009464 (rank : 27) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RAIN_MOUSE
|
||||||
| NC score | 0.009098 (rank : 28) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.008550 (rank : 29) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.006829 (rank : 30) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.005732 (rank : 31) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.001017 (rank : 32) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
ZN317_HUMAN
|
||||||
| NC score | -0.001567 (rank : 33) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PQ6, Q96PM0, Q96PM1, Q96PT2 | Gene names | ZNF317 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 317. | |||||
|
KLF10_HUMAN
|
||||||
| NC score | -0.001771 (rank : 34) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
KS6A5_HUMAN
|
||||||
| NC score | -0.003090 (rank : 35) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75582, O95316, Q96AF7 | Gene names | RPS6KA5, MSK1 | |||
|
Domain Architecture |
|
|||||
| Description | Ribosomal protein S6 kinase alpha-5 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 1) (90 kDa ribosomal protein S6 kinase 5) (RSK-like protein kinase) (RSKL). | |||||