Please be patient as the page loads
|
TCF25_MOUSE
|
||||||
| SwissProt Accessions | Q8R3L2, Q3THR8, Q3UBI7, Q3UD64, Q3UG75, Q80ZX3, Q8BR80, Q8C200, Q8C2M3, Q8C6B4, Q9CUW0, Q9ER19 | Gene names | Tcf25, D8Ertd325e, Nulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TCF25_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994612 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQ70, Q2MK75, Q9UPV3 | Gene names | TCF25, KIAA1049, NULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
TCF25_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R3L2, Q3THR8, Q3UBI7, Q3UD64, Q3UG75, Q80ZX3, Q8BR80, Q8C200, Q8C2M3, Q8C6B4, Q9CUW0, Q9ER19 | Gene names | Tcf25, D8Ertd325e, Nulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
PRPS2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 3) | NC score | 0.080922 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11908, Q15245 | Gene names | PRPS2 | |||
|
Domain Architecture |
|
|||||
| Description | Ribose-phosphate pyrophosphokinase II (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) (PPRibP). | |||||
|
PRPS2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 4) | NC score | 0.080870 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CS42, Q9CZA9 | Gene names | Prps2 | |||
|
Domain Architecture |
|
|||||
| Description | Ribose-phosphate pyrophosphokinase II (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase II) (PRS-II). | |||||
|
ZKSC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 5) | NC score | 0.004567 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.039447 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
PRPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.074914 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60891, P09329 | Gene names | PRPS1 | |||
|
Domain Architecture |
|
|||||
| Description | Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) (PPRibP). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.062222 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
PRPS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.072830 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7G0 | Gene names | Prps1 | |||
|
Domain Architecture |
|
|||||
| Description | Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PRS-I). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.052881 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
2A5B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.019803 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15173, Q13853 | Gene names | PPP2R5B | |||
|
Domain Architecture |
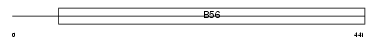
|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform (PP2A, B subunit, B' beta isoform) (PP2A, B subunit, B56 beta isoform) (PP2A, B subunit, PR61 beta isoform) (PP2A, B subunit, R5 beta isoform). | |||||
|
CN021_MOUSE
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.048088 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMC4, Q3TW87, Q8BKR9, Q8BYV4, Q8VEF9, Q9D0C6, Q9D5A8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21 homolog. | |||||
|
SGOL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.030979 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
CN021_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.042474 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U38, Q8IVF0, Q8TBS6 | Gene names | C14orf21, KIAA2021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21. | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.017666 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TRI31_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.009172 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZY9, Q53H52, Q5RI37, Q5SRJ7, Q5SRJ8, Q5SS28, Q96AK4, Q96AP8, Q99579, Q9BZY8 | Gene names | TRIM31, C6orf13, RNF | |||
|
Domain Architecture |
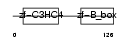
|
|||||
| Description | Tripartite motif-containing protein 31. | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.013524 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.011071 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
GLUD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.019341 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TDP6, Q9NYJ0 | Gene names | GLULD1, LGS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate--ammonia ligase domain-containing protein 1 (Lengsin) (Lens glutamine synthase-like). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.004657 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.014120 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
TCF25_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R3L2, Q3THR8, Q3UBI7, Q3UD64, Q3UG75, Q80ZX3, Q8BR80, Q8C200, Q8C2M3, Q8C6B4, Q9CUW0, Q9ER19 | Gene names | Tcf25, D8Ertd325e, Nulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
TCF25_HUMAN
|
||||||
| NC score | 0.994612 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQ70, Q2MK75, Q9UPV3 | Gene names | TCF25, KIAA1049, NULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 25 (Nuclear localized protein 1). | |||||
|
PRPS2_HUMAN
|
||||||
| NC score | 0.080922 (rank : 3) | θ value | 0.365318 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11908, Q15245 | Gene names | PRPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribose-phosphate pyrophosphokinase II (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase II) (PRS-II) (PPRibP). | |||||
|
PRPS2_MOUSE
|
||||||
| NC score | 0.080870 (rank : 4) | θ value | 0.365318 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CS42, Q9CZA9 | Gene names | Prps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribose-phosphate pyrophosphokinase II (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase II) (PRS-II). | |||||
|
PRPS1_HUMAN
|
||||||
| NC score | 0.074914 (rank : 5) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60891, P09329 | Gene names | PRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PRS-I) (PPRibP). | |||||
|
PRPS1_MOUSE
|
||||||
| NC score | 0.072830 (rank : 6) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7G0 | Gene names | Prps1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PRS-I). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.062222 (rank : 7) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.052881 (rank : 8) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
CN021_MOUSE
|
||||||
| NC score | 0.048088 (rank : 9) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMC4, Q3TW87, Q8BKR9, Q8BYV4, Q8VEF9, Q9D0C6, Q9D5A8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21 homolog. | |||||
|
CN021_HUMAN
|
||||||
| NC score | 0.042474 (rank : 10) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U38, Q8IVF0, Q8TBS6 | Gene names | C14orf21, KIAA2021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21. | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.039447 (rank : 11) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
SGOL1_MOUSE
|
||||||
| NC score | 0.030979 (rank : 12) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
2A5B_HUMAN
|
||||||
| NC score | 0.019803 (rank : 13) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15173, Q13853 | Gene names | PPP2R5B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform (PP2A, B subunit, B' beta isoform) (PP2A, B subunit, B56 beta isoform) (PP2A, B subunit, PR61 beta isoform) (PP2A, B subunit, R5 beta isoform). | |||||
|
GLUD1_HUMAN
|
||||||
| NC score | 0.019341 (rank : 14) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TDP6, Q9NYJ0 | Gene names | GLULD1, LGS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate--ammonia ligase domain-containing protein 1 (Lengsin) (Lens glutamine synthase-like). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.017666 (rank : 15) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.014120 (rank : 16) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.013524 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.011071 (rank : 18) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
TRI31_HUMAN
|
||||||
| NC score | 0.009172 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZY9, Q53H52, Q5RI37, Q5SRJ7, Q5SRJ8, Q5SS28, Q96AK4, Q96AP8, Q99579, Q9BZY8 | Gene names | TRIM31, C6orf13, RNF | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 31. | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.004657 (rank : 20) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
ZKSC1_HUMAN
|
||||||
| NC score | 0.004567 (rank : 21) | θ value | 0.365318 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||