Please be patient as the page loads
|
TAF1C_HUMAN
|
||||||
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAF1C_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
TAF1C_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.967423 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.054316 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.068953 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.069227 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
DDB2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.061070 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92466, Q76E54, Q76E55, Q76E56, Q76E57 | Gene names | DDB2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2) (DDB p48 subunit) (DDBb) (UV-damaged DNA-binding protein 2) (UV-DDB 2). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.043065 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.034649 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.032332 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
4ET_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.037382 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CN140_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.042526 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.010517 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
CORO7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.018801 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57737 | Gene names | CORO7 | |||
|
Domain Architecture |

|
|||||
| Description | Coronin-7 (70 kDa WD repeat tumor rejection antigen homolog). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.047755 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.023983 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.009245 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.024753 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
DDB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.047936 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J79 | Gene names | Ddb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.024237 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ADAM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.008744 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99965, P78326, Q9UQQ8 | Gene names | ADAM2, FTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.046732 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.025732 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
LRC24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.007797 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q50LG9 | Gene names | LRRC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 24 precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.024274 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.016254 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.013065 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
IRF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.012501 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |
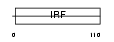
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.017086 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
HUNK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.000643 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57058 | Gene names | HUNK, MAKV | |||
|
Domain Architecture |
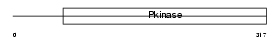
|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V) (B19). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.007168 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
LAX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.020096 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV1, Q6NSZ6, Q9NXB4 | Gene names | LAX1, LAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007184 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.007984 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
RPC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.012240 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
TPSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.015746 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15533, O15210, O15272, Q5STJ8, Q5STK6, Q5STQ5, Q5STQ6, Q66K65, Q96KK7, Q9HAN8, Q9UEE0, Q9UEE4, Q9UIZ6, Q9Y6K2 | Gene names | TAPBP, NGS17, TAPA | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein) (NGS-17). | |||||
|
TR13C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.024188 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8D0 | Gene names | Tnfrsf13c, Baffr, Bcmd, Br3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13C (B cell- activating factor receptor) (BAFF receptor) (BAFF-R) (BLyS receptor 3) (B-cell maturation defect). | |||||
|
TAF1C_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
TAF1C_MOUSE
|
||||||
| NC score | 0.967423 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.069227 (rank : 3) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.068953 (rank : 4) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
DDB2_HUMAN
|
||||||
| NC score | 0.061070 (rank : 5) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92466, Q76E54, Q76E55, Q76E56, Q76E57 | Gene names | DDB2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2) (DDB p48 subunit) (DDBb) (UV-damaged DNA-binding protein 2) (UV-DDB 2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.054316 (rank : 6) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DDB2_MOUSE
|
||||||
| NC score | 0.047936 (rank : 7) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J79 | Gene names | Ddb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.047755 (rank : 8) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.046732 (rank : 9) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.043065 (rank : 10) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CN140_MOUSE
|
||||||
| NC score | 0.042526 (rank : 11) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.037382 (rank : 12) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.034649 (rank : 13) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.032332 (rank : 14) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.025732 (rank : 15) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.024753 (rank : 16) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.024274 (rank : 17) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.024237 (rank : 18) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TR13C_MOUSE
|
||||||
| NC score | 0.024188 (rank : 19) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8D0 | Gene names | Tnfrsf13c, Baffr, Bcmd, Br3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13C (B cell- activating factor receptor) (BAFF receptor) (BAFF-R) (BLyS receptor 3) (B-cell maturation defect). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.023983 (rank : 20) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
LAX1_HUMAN
|
||||||
| NC score | 0.020096 (rank : 21) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWV1, Q6NSZ6, Q9NXB4 | Gene names | LAX1, LAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
CORO7_HUMAN
|
||||||
| NC score | 0.018801 (rank : 22) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57737 | Gene names | CORO7 | |||
|
Domain Architecture |

|
|||||
| Description | Coronin-7 (70 kDa WD repeat tumor rejection antigen homolog). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.017086 (rank : 23) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.016254 (rank : 24) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TPSN_HUMAN
|
||||||
| NC score | 0.015746 (rank : 25) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15533, O15210, O15272, Q5STJ8, Q5STK6, Q5STQ5, Q5STQ6, Q66K65, Q96KK7, Q9HAN8, Q9UEE0, Q9UEE4, Q9UIZ6, Q9Y6K2 | Gene names | TAPBP, NGS17, TAPA | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein) (NGS-17). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.013065 (rank : 26) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
IRF2_MOUSE
|
||||||
| NC score | 0.012501 (rank : 27) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
RPC1_HUMAN
|
||||||
| NC score | 0.012240 (rank : 28) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.010517 (rank : 29) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.009245 (rank : 30) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
ADAM2_HUMAN
|
||||||
| NC score | 0.008744 (rank : 31) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99965, P78326, Q9UQQ8 | Gene names | ADAM2, FTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.007984 (rank : 32) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LRC24_HUMAN
|
||||||
| NC score | 0.007797 (rank : 33) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q50LG9 | Gene names | LRRC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 24 precursor. | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.007184 (rank : 34) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.007168 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
HUNK_HUMAN
|
||||||
| NC score | 0.000643 (rank : 36) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57058 | Gene names | HUNK, MAKV | |||
|
Domain Architecture |

|
|||||
| Description | Hormonally up-regulated neu tumor-associated kinase (EC 2.7.11.1) (Serine/threonine-protein kinase MAK-V) (B19). | |||||