Please be patient as the page loads
|
SYV_HUMAN
|
||||||
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYV_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994258 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |
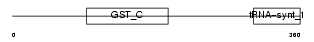
|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYIM_MOUSE
|
||||||
| θ value | 8.57503e-51 (rank : 3) | NC score | 0.760131 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_HUMAN
|
||||||
| θ value | 4.70527e-49 (rank : 4) | NC score | 0.753741 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIC_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 5) | NC score | 0.689573 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIC_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 6) | NC score | 0.688242 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
EF1G_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 7) | NC score | 0.560372 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
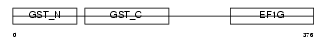
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 2.27234e-27 (rank : 8) | NC score | 0.559387 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
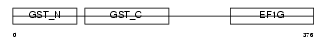
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYLM_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 9) | NC score | 0.489554 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 10) | NC score | 0.484694 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 11) | NC score | 0.249409 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYMM_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.227947 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYLC_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.248837 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.242537 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
MCA3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.166785 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.039601 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.146858 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.038352 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.006401 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MCA3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.115207 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |
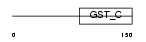
|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.060946 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.006687 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.004123 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.019009 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.019016 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.005665 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
GSTT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.062728 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |
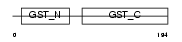
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
KS6B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.005055 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23443 | Gene names | RPS6KB1 | |||
|
Domain Architecture |
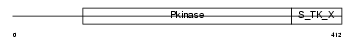
|
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K) (p70-alpha). | |||||
|
KS6B1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.005049 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSK8, Q8CHX0 | Gene names | Rps6kb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K). | |||||
|
CAH13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.005967 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N1Q1 | Gene names | CA13 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 13 (EC 4.2.1.1) (Carbonic anhydrase XIII) (Carbonate dehydratase XIII) (CA-XIII). | |||||
|
KS6B2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.004559 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBS0, O94809, Q9UEC1 | Gene names | RPS6KB2, STK14B | |||
|
Domain Architecture |
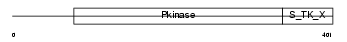
|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (p70 S6 kinase beta) (S6K-beta) (p70-beta) (S6 kinase-related kinase) (SRK) (Serine/threonine-protein kinase 14 beta). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.007580 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MICA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.006683 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PMYT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.003209 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99640, O14731, Q7LE24, Q8TCM9 | Gene names | PKMYT1, MYT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.006394 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | -0.000058 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
K0226_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.010107 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.012131 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KS6B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.003142 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.002919 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.009191 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SYM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.100086 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYV_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_MOUSE
|
||||||
| NC score | 0.994258 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYIM_MOUSE
|
||||||
| NC score | 0.760131 (rank : 3) | θ value | 8.57503e-51 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_HUMAN
|
||||||
| NC score | 0.753741 (rank : 4) | θ value | 4.70527e-49 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIC_HUMAN
|
||||||
| NC score | 0.689573 (rank : 5) | θ value | 1.01765e-35 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.688242 (rank : 6) | θ value | 3.61944e-33 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.560372 (rank : 7) | θ value | 7.80994e-28 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.559387 (rank : 8) | θ value | 2.27234e-27 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYLM_HUMAN
|
||||||
| NC score | 0.489554 (rank : 9) | θ value | 4.91457e-14 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| NC score | 0.484694 (rank : 10) | θ value | 7.09661e-13 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.249409 (rank : 11) | θ value | 1.43324e-05 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYLC_HUMAN
|
||||||
| NC score | 0.248837 (rank : 12) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| NC score | 0.242537 (rank : 13) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_HUMAN
|
||||||
| NC score | 0.227947 (rank : 14) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
MCA3_MOUSE
|
||||||
| NC score | 0.166785 (rank : 15) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.146858 (rank : 16) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
MCA3_HUMAN
|
||||||
| NC score | 0.115207 (rank : 17) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.100086 (rank : 18) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
GSTT2_HUMAN
|
||||||
| NC score | 0.062728 (rank : 19) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.060946 (rank : 20) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.039601 (rank : 21) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.038352 (rank : 22) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.019016 (rank : 23) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.019009 (rank : 24) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.012131 (rank : 25) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
K0226_MOUSE
|
||||||
| NC score | 0.010107 (rank : 26) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.009191 (rank : 27) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.007580 (rank : 28) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.006687 (rank : 29) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MICA1_HUMAN
|
||||||
| NC score | 0.006683 (rank : 30) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.006401 (rank : 31) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.006394 (rank : 32) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
CAH13_HUMAN
|
||||||
| NC score | 0.005967 (rank : 33) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N1Q1 | Gene names | CA13 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 13 (EC 4.2.1.1) (Carbonic anhydrase XIII) (Carbonate dehydratase XIII) (CA-XIII). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.005665 (rank : 34) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
KS6B1_HUMAN
|
||||||
| NC score | 0.005055 (rank : 35) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23443 | Gene names | RPS6KB1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K) (p70-alpha). | |||||
|
KS6B1_MOUSE
|
||||||
| NC score | 0.005049 (rank : 36) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSK8, Q8CHX0 | Gene names | Rps6kb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase beta-1 (EC 2.7.11.1) (Ribosomal protein S6 kinase I) (S6K) (S6K1) (70 kDa ribosomal protein S6 kinase 1) (p70 S6 kinase alpha) (p70(S6K)-alpha) (p70-S6K) (P70S6K). | |||||
|
KS6B2_HUMAN
|
||||||
| NC score | 0.004559 (rank : 37) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBS0, O94809, Q9UEC1 | Gene names | RPS6KB2, STK14B | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (p70 S6 kinase beta) (S6K-beta) (p70-beta) (S6 kinase-related kinase) (SRK) (Serine/threonine-protein kinase 14 beta). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.004123 (rank : 38) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
PMYT1_HUMAN
|
||||||
| NC score | 0.003209 (rank : 39) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99640, O14731, Q7LE24, Q8TCM9 | Gene names | PKMYT1, MYT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
KS6B2_MOUSE
|
||||||
| NC score | 0.003142 (rank : 40) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.002919 (rank : 41) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | -0.000058 (rank : 42) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||